Oxidosqualene Cyclase Knock-Down in Latex of Taraxacum koksaghyz Reduces Triterpenes in Roots and Separated Natural Rubber
Abstract
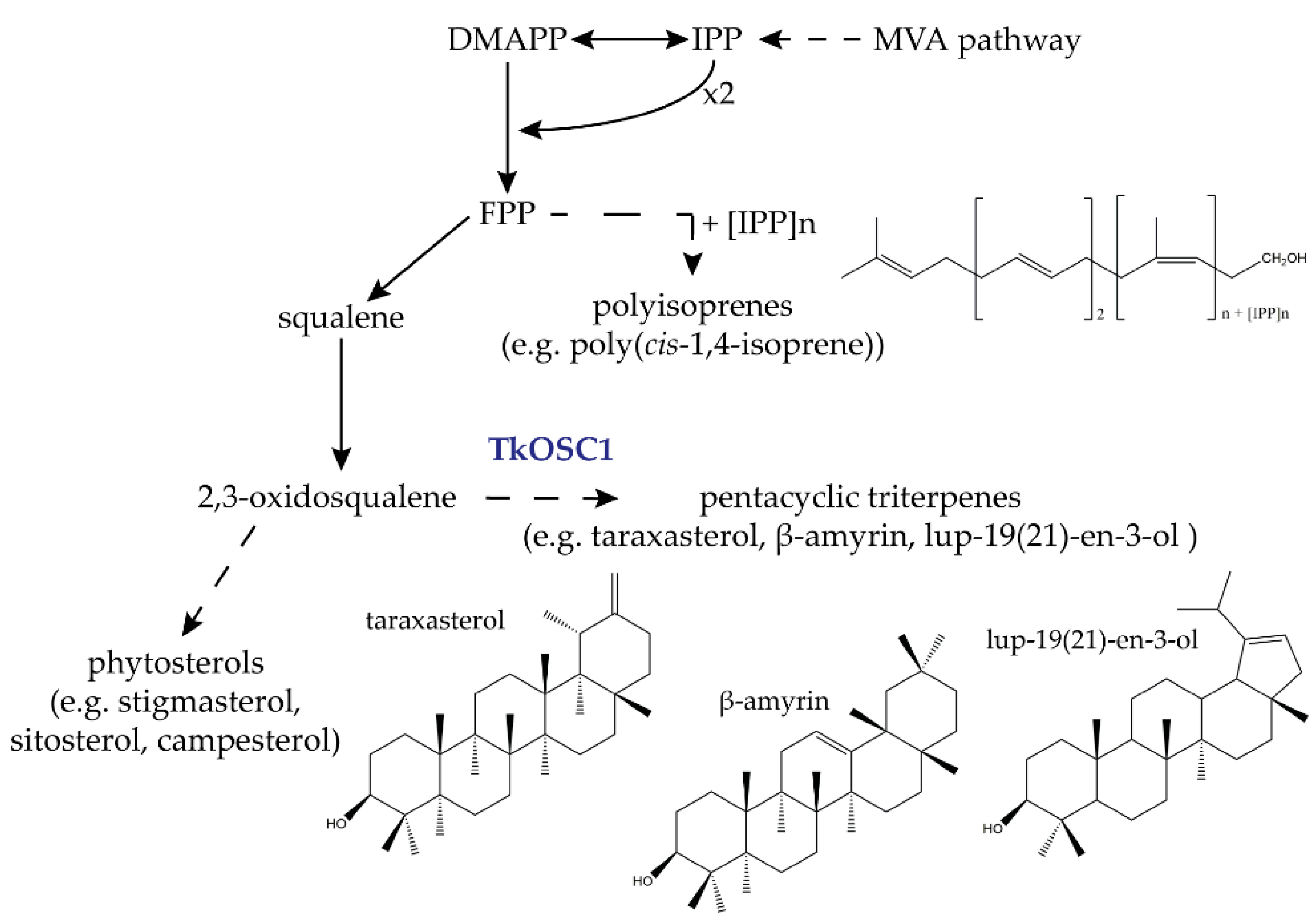
:1. Introduction
2. Results
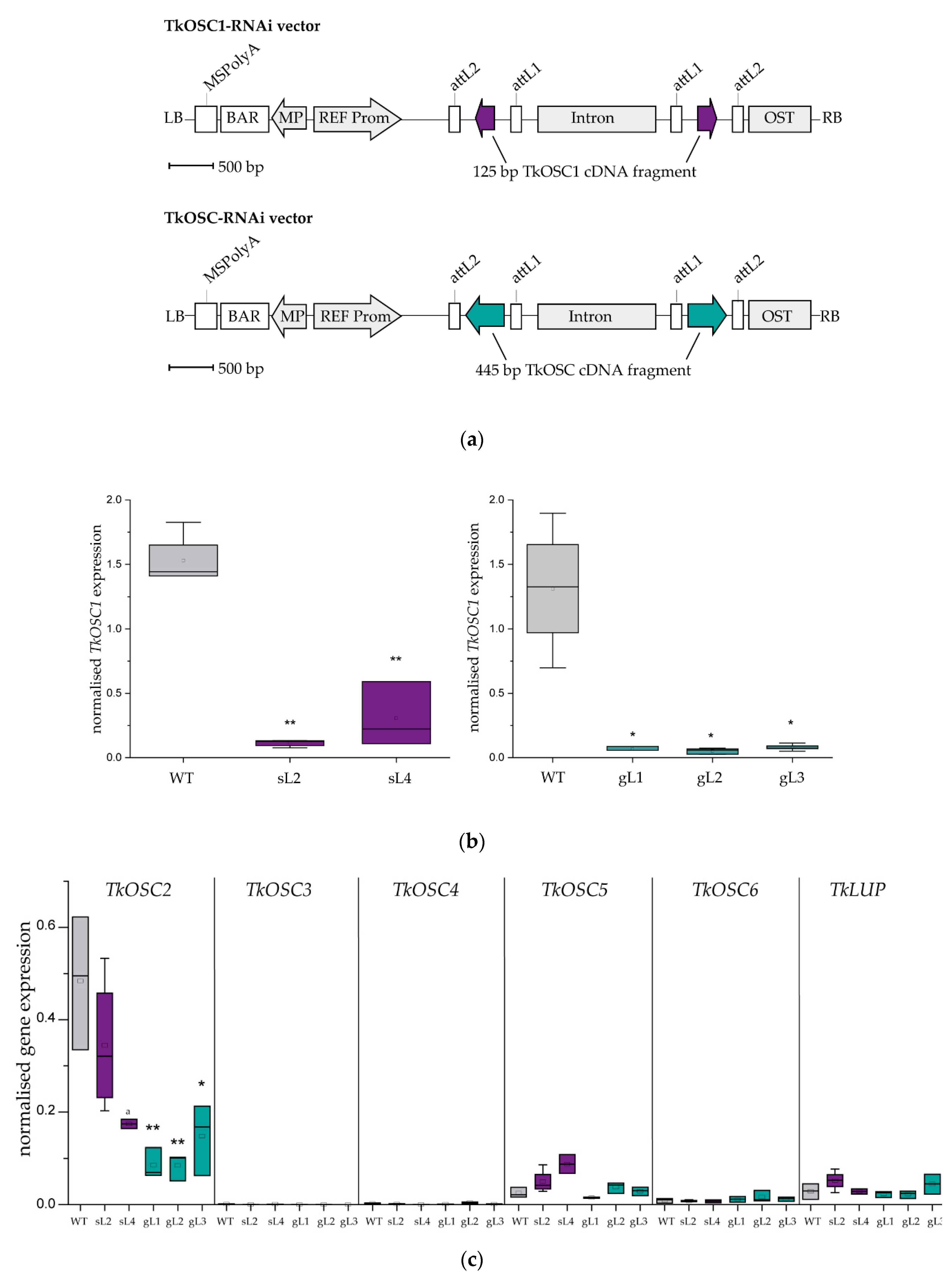
2.1. Generation of T. koksaghyz OSC RNAi Plants
2.2. Laticifer Predominant OSC Knockdown Results in Triterpene Depleted NR
2.3. Pentacyclic Triterpene Depletion has no Impact on Plant Development and Rubber Yield
3. Discussion
4. Materials and Methods
4.1. Plant Material and Cultivation Conditions
4.2. Cloning and Transformation Procedures
4.3. Quantitative Expression Analysis
4.4. Triterpene Analysis
4.5. Quantitative Determination of Poly(cis-1,4-isoprene) Levels by 1H -NMR Spectroscopy
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Furuno, T.; Kamiyama, A.; Akashi, T.; Usui, M.; Takahashi, T.; Ayabe, S. Triterpenoid constituents of tissue cultures and regenerated organs of Taraxacum officinale. Plant Tissue Cult. Lett. 1993, 10, 275–280. [Google Scholar] [CrossRef]
- Hagel, J.M.; Yeung, E.C.; Facchini, P.J. Got milk? The secret life of laticifers. Trends Plant Sci. 2008, 13, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Mooibroek, H.; Cornish, K. Alternative sources of natural rubber. Appl. Microbiol. Biotechnol. 2000, 53, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Schulze Gronover, C.; Wahler, D.; Prüfer, D. Natural rubber biosynthesis and physic-chemical studies on plant derived latex. In Biotechnology of Biopolymers; Magdy Elnashar, Ed.; IntechOpen: London, UK, 2011; pp. 75–88. [Google Scholar]
- Krotkov, G. A review of literature on Taraxacum kok-saghyz Rod. Bot. Rev. 1945, 9, 417–461. [Google Scholar] [CrossRef]
- Nor, H.M.; Ebdon, J.R. Telechelic liquid natural rubber: A review. Prog. Polym. Sci. 1998, 23, 143–177. [Google Scholar] [CrossRef]
- Sakdapipanich, J.T. Structural characterization of natural rubber based on recent evidence from selective enzymatic treatments. J. Biosci. Bioeng. 2007, 103, 287–292. [Google Scholar] [CrossRef]
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.-L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 2016, 34, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Surmacz, L.; Swiezewska, E. Polyisoprenoids-Secondary metabolites or physiologically important superlipids? Biochem. Biophys. Res. Commun. 2011, 407, 627–632. [Google Scholar] [CrossRef]
- Abe, I. Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 2007, 24, 1311–1331. [Google Scholar] [CrossRef]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef]
- Xu, R.; Fazio, G.C.; Matsuda, S.P.T. On the origins of triterpenoid skeletal diversity. Phytochemistry 2004, 65, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.A. Plant sterols and the membrane environment. Trends Plant Sci. 1998, 3, 170–175. [Google Scholar] [CrossRef]
- Augustin, J.M.; Kuzina, V.; Andersen, S.B.; Bak, S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 2011, 72, 435–457. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A.; Goss, R.J.; Field, R.A. The saponins: Polar isoprenoids with important and diverse biological activities. Nat. Prod. Rep. 2011, 28, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Papadopoulou, K.K.; Osbourn, A. Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pütter, K.M.; van Deenen, N.; Müller, B.; Fuchs, L.; Vorwerk, K.; Unland, K.; Bröker, J.N.; Scherer, E.; Huber, C.; Eisenreich, W.; et al. The enzymes OSC1 and CYP716A263 produce a high variety of triterpenoids in the latex of Taraxacum koksaghyz. Sci. Rep. 2019, 9, 5942. [Google Scholar] [CrossRef]
- Bröker, J.N.; Müller, B.; van Deenen, N.; Prüfer, D.; Schulze Gronover, C. Upregulating the mevalonate pathway and repressing sterol synthesis in Saccharomyces cerevisiae enhances the production of triterpenes. Appl. Microbiol. Biotechnol. 2018, 102, 6923–6934. [Google Scholar] [CrossRef] [PubMed]
- Laibach, N.; Hillebrand, A.; Twyman, R.M.; Prüfer, D.; Schulze Gronover, C. Identification of a Taraxacum brevicorniculatum rubber elongation factor protein that is localized on rubber particles and promotes rubber biosynthesis. Plant J. 2015, 82, 609–620. [Google Scholar] [CrossRef]
- Han, J.Y.; Kwon, Y.S.; Yang, D.C.; Jung, Y.R.; Choi, Y.E. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 2006, 47, 1653–1662. [Google Scholar] [CrossRef]
- Yang, J.L.; Hu, Z.F.; Zhang, T.T.; Gu, A.D.; Gong, T.; Zhu, P. Progress on the Studies of the Key Enzymes of Ginsenoside Biosynthesis. Molecules 2018, 23, 589. [Google Scholar] [CrossRef]
- Mishra, S.; Bansal, S.; Mishra, B.; Sangwan, B.S.; Asha; Jadaun, J.S.; Sangwan, N.S. RNAi and Homologous Over-Expression Based Functional Approaches Reveal Triterpenoid Synthase Gene-Cycloartenol Synthase Is Involved in Downstream Withanolide Biosynthesis in Withania somnifera. PLoS ONE 2016, 11, e0149691. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ge, F.; Sun, Y.; Liu, D.; Chen, C. Strengthening Triterpene Saponins Biosynthesis by Over-Expression of Farnesyl Pyrophosphate Synthase Gene and RNA Interference of Cycloartenol Synthase Gene in Panax notoginseng Cells. Molecules 2017, 22, 581. [Google Scholar] [CrossRef] [PubMed]
- Takagi, K.; Nishizawa, K.; Hirose, A.; Kita, A.; Ishomoto, M. Manipulation of saponin biosynthesis by RNA interference-mediated silencing of β-amyrin synthase gene expression in soybean. Plant Cell Rep. 2011, 30, 1835–1846. [Google Scholar] [CrossRef] [PubMed]
- Pütter, K.M.; van Deenen, N.; Unland, K.; Prüfer, D.; Schulze Gronover, C. Isoprenoid biosynthesis in dandelion latex is enhanced by the overexpression of three key enzymes involved in the mevalonate pathway. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Epping, J.; van Deenen, N.; Niephaus, E.; Stolze, A.; Fricke, J.; Huber, C.; Eisenreich, W.; Twyman, R.M.; Prüfer, D.; Schulze Gronover, C. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 2015, 1, 15048. [Google Scholar] [CrossRef]
- Stolze, A.; Wanke, A.; van Deenen, N.; Geyer, R.; Prüfer, D.; Schulze Gronover, C. Development of rubber-enriched dandelion varieties by metabolic engineering of the inulin pathway. Plant Biotechnol. J. 2017, 15, 740–753. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Lenders, M.; Hillebrand, A.; van Deenen, N.; Munt, O.; Reichelt, R.; Eisenreich, W.; Fischer, R.; Prüfer, D.; Schulze Gronover, C. Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochem. 2010, 11, 11. [Google Scholar] [CrossRef]
Sample Availability: Not available. |


| mg g−1 NR | WT | TkOSC1-RNAi sL2/sL4 | TkOSC-RNAigL1/gL2 |
|---|---|---|---|
| precursor | 1.0 () | 3.5 ()* | 8.6 () ** |
| sterols | 1.2 () | 2.1 () | 1.4 () |
| pentacyclic triterpenes | 56.6 () | 33.4 ()* | 16.0 () ** |
| total triterpenes | 58.9 () | 39.1 ()* | 26.1 () ** |
| pentacyclic triterpene reduction | −41.0% | −71.6% | |
| total triterpene reduction | −33.6% | −55.6% |
| mg g−1 NR | WT | gL2 | gL3 |
|---|---|---|---|
| precursor | 1.0 () | 6.8 () ** | 3.3 () ** |
| sterols | 0.9 () | 0.9 () | 0.9 () |
| pentacyclic triterpenes | 57.3 () | 11.8 ()** | 15.4 () ** |
| total triterpenes | 59.3 () | 19.5 () ** | 19.6 () ** |
| pentacyclic triterpene reduction | −79.5% | −73.1% | |
| total triterpene reduction | −67.0% | −66.9% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Deenen, N.; Unland, K.; Prüfer, D.; Schulze Gronover, C. Oxidosqualene Cyclase Knock-Down in Latex of Taraxacum koksaghyz Reduces Triterpenes in Roots and Separated Natural Rubber. Molecules 2019, 24, 2703. https://doi.org/10.3390/molecules24152703
van Deenen N, Unland K, Prüfer D, Schulze Gronover C. Oxidosqualene Cyclase Knock-Down in Latex of Taraxacum koksaghyz Reduces Triterpenes in Roots and Separated Natural Rubber. Molecules. 2019; 24(15):2703. https://doi.org/10.3390/molecules24152703
Chicago/Turabian Stylevan Deenen, Nicole, Kristina Unland, Dirk Prüfer, and Christian Schulze Gronover. 2019. "Oxidosqualene Cyclase Knock-Down in Latex of Taraxacum koksaghyz Reduces Triterpenes in Roots and Separated Natural Rubber" Molecules 24, no. 15: 2703. https://doi.org/10.3390/molecules24152703





