Antiproliferative Benzoindazolequinones as Potential Cyclooxygenase-2 Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. In Silico Virtual Screening for Potential Antineoplastic Targets of BIZQs
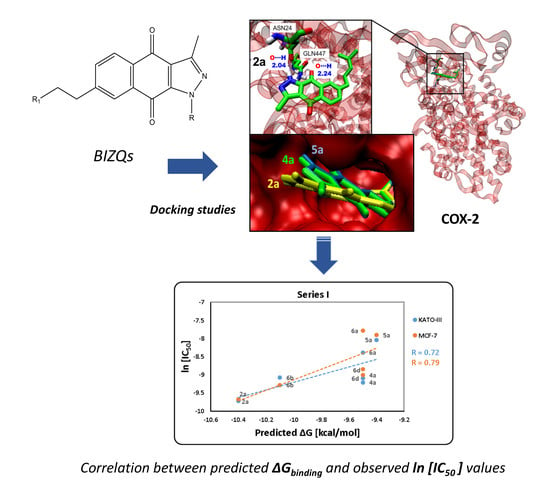
2.3. Binding Site and Docking of BIZQs in COX-2
2.4. In Vitro Cytotoxicity Results and their Correlation with Physicochemical and Pharmaco-Toxicological Parameters of BIZQs
2.5. In Silico ADME Studies
3. Materials and Methods
3.1. Chemistry
3.2. Computational Details
3.2.1. Ligand Preparation
3.2.2. In Silico ADME Prediction
3.2.3. Macromolecules Selection and Retrieve
3.2.4. Molecular Docking of Ligand-Protein Interaction
3.3. Biological Activity
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Rosen, J.; Mangiameli, D.; Libutti, S.K. Cancer development and progression. Adv. Exp. Med. Biol. 2007, 593, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Florea, A.-M.; Büsselberg, D. Cisplatin as an Anti-Tumor Drug: Cellular Mechanisms of Activity, Drug Resistance and Induced Side Effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Khadjavi, A.; Stura, I.; Prato, M.; Minero, V.G.; Panariti, A.; Rivolta, I.; Gulino, G.R.; Bessone, F.; Giribaldi, G.; Quaglino, E.; et al. ‘In Vitro-’, ‘In Vivo’ and ‘In Silico’ Investigation of the Anticancer Effectiveness of Oxygen-Loaded Chitosan-Shelled Nanodroplets as Potential Drug Vector. Pharm. Res. 2018, 35, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Saibu, M.; Sagar, S.; Green, I.; Ameer, F.; Meyer, M. Evaluating the cytotoxic effects of novel quinone compounds. Anticancer Res. 2014, 34, 4077–4086. [Google Scholar] [PubMed]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, D.D.; Chapolikar, A.D.; Devkate, C.G.; Warad, K.D.; Tayade, A.P.; Pawar, R.P.; Domb, A.J. Synthesis of indazole motifs and their medicinal importance: An overview. Eur. J. Med. Chem. 2015, 90, 707–731. [Google Scholar] [CrossRef] [PubMed]
- Thangadurai, A.; Minu, M.; Wakode, S.; Agrawal, S.; Narasimhan, B. Indazole: A medicinally important heterocyclic moiety. Med. Chem. Res. 2012, 21, 1509–1523. [Google Scholar] [CrossRef]
- Fieser, L.F.; Peters, M.A. The addition of diazomethane and some of its derivatives to alpha-naphthoquinone. J. Am. Chem. Soc. 1931, 53, 4080–4093. [Google Scholar] [CrossRef]
- Molinari, A.; Oliva, A.; Arismendi, M.; Imbarack, E.; Gálvez, C.; Maldonado, J.; San Feliciano, A. The Synthesis of Some Fused Pyrazolo-1,4-Naphthoquinones. J. Heterocycl. Chem. 2015, 52, 620–622. [Google Scholar] [CrossRef]
- Conway, G.A.; Loeffler, L.J.; Hall, I.H. Synthesis and Antitumor Evaluation of Seleted 5,6-Disubstituted 1(2) H-Indazole-4,7-diones. J. Med. Chem. 1983, 26, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Dong, Y.; Gao, J.; Gong, M.; Zhang, X.; Kong, W.; Li, Y.; Zeng, Y.; Si, D.; Wei, Z.; et al. Aspartate-modified doxorubicin on its N-terminal increases drug accumulation in LAT1-overexpressing tumors. Cancer Sci. 2015, 106, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Molinari, A.; Oliva, A.; Arismendi-Macuer, M.; Guzmán, L.; Fuentealba, M.; Knox, M.; Vinet, R.; San Feliciano, A. New 1H-benzo[f]indazole-4,9-diones conjugated with C-protected amino acids and other derivatives: Synthesis and in vitro antiproliferative evaluation. Molecules 2015, 20, 21924–21938. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, A.R. Helicobacter, Inflammation, and Gastric Cancer. Curr. Pathobiol. Rep. 2013, 1, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Huang, C.-Z. Mitogen-activated protein kinase signaling pathway and invasion and metastasis of gastric cancer. World J. Gastroenterol. 2015, 21, 11673–11679. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, D.A.J.; Negm, O.H.; Alabdullah, M.L.; Mirza, S.; Hamed, M.R.; Band, V.; Green, A.R.; Ellis, I.O.; Rakha, E.A. Clinicopathological and prognostic significance of mitogen-activated protein kinases (MAPK) in breast cancers. Breast Cancer Res. Treat. 2016, 159, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, O. Targeting vascular endothelial growth factor (VEGF) pathway in gastric cancer: Preclinical and clinical aspects. Crit. Rev. Oncol. Hematol. 2015, 93, 18–27. [Google Scholar] [CrossRef]
- Li, T.; Yu, J.; Luo, X.; Ren, W.; Zhang, Y.; Cao, B. VEGFR-2 as a novel predictor of survival in gastric cancer: A systematic review and meta-analysis. Pathol. Res. Pract. 2018, 214, 560–564. [Google Scholar] [CrossRef]
- Khanna, P.; Chua, P.J.; Bay, B.H.; Baeg, G.H. The JAK/STAT signaling cascade in gastric carcinoma (Review). Int. J. Oncol. 2015, 47, 1617–1626. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.Y.; Guo, J.L.; Jiang, Z.N.; Xie, S.D.; Shen, J.G.; Shen, J.Y.; Wang, L.B. Prognostic role of estrogen receptor α and estrogen receptor β in gastric cancer. Ann. Surg. Oncol. 2010, 17, 2503–2509. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, S.C.; Couse, J.F.; Korach, K.S. Estrogen receptor knockout mice: What their phenotypes reveal about mechanisms of estrogen action. Breast Cancer Res. 2000, 2, 345–352. [Google Scholar] [CrossRef]
- Izzo, J.G.; Ajani, J.A. Thinking In and Out of the Box When It Comes to Gastric Cancer and Cyclooxygenase-2. J. Clin. Oncol. 2007, 25, 4865–4867. [Google Scholar] [CrossRef] [PubMed]
- Sobolewski, C.; Cerella, C.; Dicato, M.; Ghibelli, L.; Diederich, M. The role of cyclooxygenase-2 in cell proliferation and cell death in human malignancies. Int. J. Cell Biol. 2010, 215158. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Berry, J.A.; Shoher, A.; Ramakrishnan, V.; Lucci, A. COX-2 overexpression increases motility and invasion of breast cancer cells. Int. J. Oncol. 2005, 26, 1393–1399. [Google Scholar] [CrossRef]
- Kato, M.; Asaka, M. Recent Knowledge of the Relationship Between Helicobacter pylori and Gastric Cancer and Recent Progress of Gastroendoscopic Diagnosis and Treatment for Gastric Cancer. Jpn. J. Clin. Oncol. 2010, 40, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Svrcek, M.; El-Bchiri, J.; Chalastanis, A.; Capel, E.; Dumont, S.; Buhard, O.; Oliveira, C.; Seruca, R.; Bossard, C.; Mosnier, J.F.; et al. Specific Clinical and Biological Features Characterize Inflammatory Bowel Disease—Associated Colorectal Cancers Showing Microsatellite Instability. J. Clin. Oncol. 2007, 25, 4231–4238. [Google Scholar] [CrossRef]
- Actelion Pharmaceuticals Ltd. DataWarrior: A Free Cheminformatics Program for Data Visualization and Analysis. Available online: www.openmolecules.org/datawarrior/ (accessed on 6 February 2019).
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 12, 1097–1105. [Google Scholar] [CrossRef]
- Bhattarai, P.; Hameed, S.; Dai, Z. Recent advances in anti-angiogenic nanomedicines for cancer therapy. Nanoscale 2018, 10, 5393–5423. [Google Scholar] [CrossRef]
- Kieran, M.W.; Kalluri, R.; Cho, Y.J. The VEGF pathway in cancer and disease: Responses, resistance, and the path forward. Cold Spring Harb. Perspect Med. 2012, 2, a006593. [Google Scholar] [CrossRef]
- Hu, H.; Han, T.; Zhuo, M.; Wu, L.L.; Yuan, C.; Wu, L.; Lei, W.; Jiao, F.; Wang, L.W. Elevated COX-2 Expression Promotes Angiogenesis Through EGFR/p38-MAPK/Sp1-Dependent Signalling in Pancreatic Cancer. Sci. Rep. 2017, 7, 470. [Google Scholar] [CrossRef]
- Xu, L.; Stevens, J.; Hilton, M.B.; Seaman, S.; Conrads, T.P.; Veenstra, T.D.; Logsdon, D.; Morris, H.; Swing, D.A.; Patel, N.L.; et al. COX-2 inhibition potentiates antiangiogenic cancer therapy and prevents metastasis in preclinical models. Sci. Transl. Med. 2014, 6, 242ra84. [Google Scholar] [CrossRef]
- ACD/Labs.com:: Your Partner in Chemistry Software for Analytical and Chemical Knowledge Management, Chemical Nomenclature, and In-Silico PhysChem and ADME-Tox. Available online: https://www.acdlabs.com/ (accessed on 7 July 2018).
- Zarghi, A.; Arfaei, S. Selective COX-2 Inhibitors: A Review of Their Structure-Activity Relationships. Iran J. Pharm. Res. 2011, 10, 655–683. [Google Scholar] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 03; Gaussian Inc.: Wallingford, CT, USA, 2004. [Google Scholar]
- Schrödinger, LLC, New York, NY. Available online: https://www.schrodinger.com/ (accessed on 7 July 2018).
- Jorgensen, W.L.; Duffy, E.M. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002, 54, 355–366. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |






| Compd. | PDB Entries | C avge. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1DLS | 1GJO | 2OJG | 2QTU | 2ZZ0 | 3ERT | 3LN1 | 3RXH | 3VHE | 4AN3 | 4EHZ | 5GWK | ||
| Series Ia | |||||||||||||
| 2a | −7.2 | −8.5 | −9.4 | −9.8 | −8.4 | −8.2 | −10.4 | −9.1 | −10.2 | −9.3 | −9.5 | −8.3 | −9.03 |
| 3a | −8.9 | −8.5 | −9.6 | −8.8 | −8.6 | −7.9 | −10.0 | −9.1 | −10.9 | −9.3 | −9.4 | −8.3 | −9.11 |
| 4a | −7.7 | −8.3 | −8.8 | −8.6 | −8.5 | −7.9 | −9.5 | −8.2 | −9.9 | −8.4 | −8.8 | −7.2 | −8.48 |
| 5a | −8.0 | −8.3 | −9.2 | −9.0 | −8.8 | −7.2 | −9.4 | −8.6 | −8.4 | −8.8 | −9.0 | −7.4 | −8.51 |
| Series Ib | |||||||||||||
| 6a | −8.0 | −8.4 | −9.1 | −8.2 | −9.3 | −7.5 | −9.5 | −8.2 | −8.4 | −9.0 | −9.1 | −6.9 | −8.47 |
| 6b | −9.0 | −8.5 | −8.8 | −8.4 | −9.4 | −7.8 | −10.1 | −8.2 | −8.9 | −9.0 | −9.2 | −7.5 | −8.72 |
| 6c | −10.0 | −9.8 | −9.4 | −9.0 | −9.5 | −8.5 | −10.4 | −8.4 | −8.8 | −9.3 | −10.3 | −8.5 | −9.33 |
| 6d | −8.8 | −8.6 | −8.9 | −7.8 | −8.7 | −8.0 | −9.5 | −7.9 | −7.9 | −8.8 | −9.0 | −6.7 | −8.38 |
| Series IIa | |||||||||||||
| 2b | −7.6 | −8.1 | −9.2 | −8.5 | −8.2 | −7.9 | −9.8 | −8.7 | −9.4 | −8.8 | −8.7 | −7.5 | −8.53 |
| 3b | −8.6 | −8.3 | −9.1 | −8.0 | −8.5 | −7.7 | −9.8 | −8.6 | −9.1 | −9.0 | −9.3 | −7.4 | −8.62 |
| 4b | −7.6 | −8.0 | −8.9 | −7.6 | −7.9 | −7.5 | −8.7 | −7.9 | −7.9 | −8.2 | −8.0 | −7.4 | −7.97 |
| 5b | −8.0 | −8.0 | −9.2 | −7.9 | −8.3 | −7.8 | −9.3 | −8.2 | −8.3 | −8.6 | −8.9 | −6.3 | −8.23 |
| Series IIb | |||||||||||||
| 6e | −8.1 | −8.1 | −9.2 | −7.9 | −8.6 | −7.7 | −9.4 | −8.3 | −8.9 | −8.9 | −8.9 | −7.2 | −8.43 |
| 6f | −8.3 | −8.3 | −9.3 | −7.8 | −8.8 | −7.8 | −9.7 | −8.3 | −8.3 | −8.6 | −8.8 | −7.4 | −8.45 |
| 6g | −9.0 | −9.4 | −9.7 | −9.0 | −9.8 | −9.5 | −10.1 | −8.7 | −8.5 | −9.2 | −10.0 | −8.0 | −9.24 |
| 6h | −8.6 | −8.4 | −8.9 | −7.8 | −8.8 | −8.1 | −8.8 | −8.0 | −8.0 | −8.0 | −8.5 | −7.4 | −8.28 |
| Series IIIa | |||||||||||||
| 2c | −8.5 | −8.4 | −9.3 | −8.5 | −8.9 | −7.9 | −10.0 | −8.3 | −9.6 | −9.2 | −9.2 | −7.5 | −8.78 |
| 3c | −8.9 | −8.4 | −9.5 | −8.2 | −9.0 | −8.0 | −9.5 | −8.5 | −9.2 | −9.6 | −9.4 | −7.7 | −8.83 |
| 4c | −8.1 | −8.8 | −8.8 | −7.2 | −8.1 | −7.2 | −9.3 | −7.9 | −8.3 | −8.4 | −8.8 | −6.7 | −8.13 |
| 5c | −8.2 | −8.4 | −9.1 | −7.8 | −8.4 | −7.6 | −9.7 | −8.3 | −8.6 | −8.8 | −8.9 | −6.9 | −8.39 |
| Series IIIb | |||||||||||||
| 6i | −8.7 | −8.4 | −9.0 | −7.9 | −9.1 | −8.1 | −9.7 | −8.2 | −9.2 | −8.7 | −9.0 | −7.3 | −8.57 |
| 6j | −8.5 | −8.3 | −8.8 | −8.1 | −9.5 | −8.2 | −9.8 | −8.3 | −9.2 | −8.9 | −8.7 | −7.6 | −8.66 |
| 6k | −9.5 | −9.5 | −9.5 | −8.9 | −9.6 | −10.0 | −10.2 | −9.0 | −8.8 | −9.1 | −10.4 | −8.5 | −9.42 |
| 6m | −9.0 | −8.6 | −8.7 | −7.9 | −9.2 | −8.4 | −9.7 | −7.8 | −8.7 | −8.7 | −8.9 | −6.8 | −8.60 |
| P avge. | −8.50 | −8.51 | −9.14 | −7.9 | −8.83 | −8.02 | −9.68 | −8.40 | −8.55 | −8.86 | −9.12 | −7.43 | |
| BIZQ | H-Bonds and Hydrophobic Contacts in the Binding Site * |
|---|---|
| Series Ia | |
| 2a | Cys21, Asn24, Asn28, Arg29, Gly30, Glu31, Cys32//Tyr116, Leu138, Pro139//Glu451, Gln447, Lys454, Arg455 |
| 3a | His193, Ph196, Lys197, Thr198, His200//Asn368, Tyr371, His372, Trp373, His374 |
| 4a | Asn24, Asn28, Arg29, Gly30, Cys32//Tyr116, Gly121, Leu138, Pro139//Lys454, Arg455 |
| 5a | Asn24, Gln27, Asn28, Arg29, Gly30, Glu31,Cys32//Tyr116, Leu138, Pro139//Gln447, Lys454, Glu451, Arg455 |
| Series Ib | |
| 6a | Asn19, Cys21, Cys22, Asn24, Cys26, Arg29, Gly30, Cys32//Tyr116, Leu138, Pro139, Pro140, Val141, Ala142//Gln447, Arg455 |
| 6b | Cys21, Asn24, Gln27, Asn28, Arg29, Gly30, Glu31, Cys32//Gly121, Leu138, Pro139//Gln447, Glu451, Lys454, Arg455 |
| 6c | Asn19, Cys21, Cys22, Asn24, Cys26, Arg29, Gly30, Glu31, Cys32//Tyr116, Gly121, Tyr122, Leu138, Pro139, Pro140, Val141, Ala142//Arg455 |
| 6d | Asn19, Cys21, Cys22, Asn24, Cys26, Arg29, Cys32//Tyr116, Gly121, Leu138, Pro139, Pro140, Val141, Ala142//Lys454, Arg455 |
| Series IIa | |
| 2b | Cys21, Cys22, Asn24, Arg29, Cys32, Gln44//Asp111, Tyr116, Gly121, Ala137, Leu138, Pro139, Pro140, Ala142, Cys145//Gln447, Arg455 |
| 3b | Asn19, Cys21, Asn24, Cys32, His119, Tyr120, Gly121,//Tyr122, Pro139, Pro140, Val141, Ala142//Gln447 |
| 4b | Cys21, Asn24, Cys32//His119, Tyr120, Gln121, Tyr122, Pro139, Pro140, Val141, Ala142//Gln447 |
| 5b | Cys21, Asn24, Cys26, Gly27, Asn28, Arg29, Gly30, Glu31, Cys32//Tyr116, Gly121, Leu138, Pro139//Gln447, Glu451, Lys454, Arg455 |
| Series IIb | |
| 6e | Asn19, Cys21, Asn24, Cys26, Arg29, Gly30, Glu31, Cys32//Tyr116, Gly121, Tyr122, Leu138, Pro139, Ala142//Gln447, Glu451 |
| 6f | Asn19, Cys21, Asn24, Cys26, Arg29, Gly30, Glu31, Cys32, Met33, Ser34//Tyr116, Gly121, Tyr122, Leu138, Pro139//Gln447, Glu451 |
| 6g | Asn19, Cys21, Cys22, Asn24, Cys32//Val118, His119, Gly121, Tyr122, Pro139, Pro140, Val141, Ala142, Asp143, Cys145 |
| 6h | Asn19, Cys21, Asn24, Cys26, Arg29, Gly30, Glu31, Cys32//Tyr116, His119, Gly121, Tyr122, Leu138, Pro139, Pro140, Ala142//Gln447, Glu451 |
| Series IIIa | |
| 2c | Cys21, Cys22, Asn24, Asn28, Arg29, Gly30, Glu31, Cys32//Tyr116, Gly121, Leu138, Pro139, Val141, Ala142//Gln447, Glu451, Lys454, Arg455 |
| 3c | Gln178//Asp333, Gln336, His337, Tyr341//Ile550, Ser565, Phe566, Asn567 |
| 4c | Arg29, Cys21, Asn24, Gln27, Asn28, Gly30, Glu31, Cys32//Tyr116, Gly121, Leu138, Pro139, Pro140, Val141, Ala142//Glu451, Lys454, Arg455 |
| 5c | Arg29, Cys21, Asn24, Gln27, Asn28, Gly30, Glu31//Tyr116, Gly121, Leu138, Pro139, Pro140, Val141, Ala142//Gln447, Glu451, Lys454, Arg455 |
| Series IIIb | |
| 6i | Asn19, Cys21, Gln27, Asn28, Arg29, Gly30, Glu31, Cys32//Tyr116, Gly121, Tyr122, Leu138, Pro139, Pro140, Ala142//Glu451, Lys454, Arg455 |
| 6j | Asn28, Arg29, Arg46, Thr47, Phe49//Tyr108, Leu138//Glu451, Lys454, Arg455 |
| 6k | Asn19, Arg29, Cys21, Asn24, Gln27, Asn28, Gly30, Glu31, Cys32//Tyr116, Gly121, Tyr122, Leu138, Pro139, Ala142, Asp143//Glu451, Lys454, Arg455 |
| 6m | Asn19, Cys21, Asn24, Gln27, Asn28, Arg29, Gly30, Glu31, Cys32, Met33, Ser34//Tyr116, Gly121, Tyr122, Leu138, Pro139, Ala142//Glu451, Lys454, Arg455 |
| BIZQ | In Vitro pIC50 a | ∆Gbin (kcal/mol) | Predicted Parameters | D-like | Toxicity Risks b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KATO-III | MCF-7 | COX-2 | 3avge.c | pKa1/pKa2 d,e | cLogP f | M | T | R | I | ||
| Subseries a | |||||||||||
| 2a | 4.23 ± 0.22 | 4.20 ± 0.17 | −10.4 | −9.78 | 8.86/--- | 3.856 | 00.716 | n | n | n | n |
| 2b | 4.59 ± 0.18 | 4.56 ± 0.25 | −9.8 | −9.23 | 14.94/--- | 3.354 | 22.137 | n | n | n | n |
| 2c | 4.48 ± 0.26 | 4.53 ± 0.14 | −10.0 | −9.50 | ---/--- | 3.838 | 33.050 | n | n | n | n |
| 3a | 3.50 ± 0.19 | 3.36 ± 0.18 | −10.0 | −9.67 | 8.86/--- | 2.425 | 00.351 | l | l | h | n |
| 3b | 3.40 ± 0.16 | 3.38 ± 0.13 | −9.8 | −9.40 | 14.94/--- | 1.923 | 11.691 | l | l | h | n |
| 3c | 3.49 ± 0.17 | 3.41 ± 0.10 | −9.5 | −9.47 | ---/--- | 2.408 | 22.564 | l | l | h | n |
| 4a | 4.00 ± 0.13 | 3.91 ± 0.21 | −9.5 | −9.03 | 8.86/--- | 1.454 | 00.087 | h | n | n | h |
| 4b | 4.20 ± 0.19 | 4.36 ± 0.16 | −8.7 | −8.53 | 14.94/--- | 0.952 | 11.479 | h | n | n | h |
| 4c | 4.22 ±0.13 | 4.48 ± 0.21 | −9.3 | −8.97 | ---/--- | 1.436 | 22.196 | h | n | n | h |
| 5a | 3.49 ± 0.12 | 3.43 ± 0.13 | −9.4 g | −9.20 | 3.17/8.86 | 1.317 | 22.104 | n | n | n | n |
| 5b | 3.47 ± 0.10 | 3.48 ± 0.11 | −9.3 g | −9.13 | 3.15/14.94 | 0.815 | 33.465 | n | n | n | n |
| 5c | 3.79 ± 0.15 | 3.61 ± 0.18 | −9.7 g | −9.23 | 3.17 | 1.299 | 44.378 | n | n | n | n |
| Subseries b | |||||||||||
| 6a | 3.64 ± 0.19 | 3.38 ± 0.20 | −9.5 | −9.23 | 8.86/11.52 | 0.828 | 00.103 | n | n | n | n |
| 6e | 3.51 ± 0.16 | 3.54 ± 0.12 | −9.4 | −9.17 | 11.31/14.94 | 0.326 | 11.123 | n | n | n | n |
| 6i | 3.68 ± 0.15 | 3.59 ± 0.15 | −9.7 | −9.23 | 11.32/--- | 0.811 | 22.001 | n | n | n | n |
| 6b | 3.94 ± 0.13 | 4.03 ± 0.14 | −10.1 | −9.37 | 8.86/11.41 | 1.187 | −1.153 | n | n | n | n |
| 6f | 4.36 ± 0.19 | 4.20 ± 0.24 | −9.7 | −9.27 | 11.20/14.94 | 0.685 | 0.042 | n | n | n | n |
| 6j | 4.27 ± 0.26 | 4.28 ± 0.14 | −9.8 | −9.10 | 11.21/--- | 1.170 | 0.867 | n | n | n | n |
| 6c | 3.90 ± 0.21 | 3.81 ± 0.18 | −10.4 | −10.03 | 8.86/11.50 | 2.629 | 2.272 | n | n | n | n |
| 6g | 4.42 ± 0.22 | 4.41 ± 0.22 | −10.1 | −9.93 | 11.32/14.94 | 2.127 | 3.478 | n | n | n | n |
| 6k | 4.46 ± 0.14 | 4.45 ± 0.27 | −10.2 | −10.03 | 11.33/--- | 2.612 | 4.317 | n | n | n | n |
| 6d | 3.95 ± 0.21 | 3.84 ± 0.16 | −9.5 | −9.13 | 8.86/11.39 | 1.248 | −3.119 | n | n | n | n |
| 6h | 4.28 ± 0.21 | 4.06 ± 0.12 | −8.8 | −8.73 | 11.22/14.94 | 0.746 | −1.914 | n | n | n | n |
| 6m | 3.96 ± 0.23 | 4.00 ± 0.12 | −9.7 | −9.10 | 11.22/--- | 1.230 | −1.060 | n | n | n | n |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molinari, A.; Oliva, A.; Arismendi-Macuer, M.; Guzmán, L.; Acevedo, W.; Aguayo, D.; Vinet, R.; San Feliciano, A. Antiproliferative Benzoindazolequinones as Potential Cyclooxygenase-2 Inhibitors. Molecules 2019, 24, 2261. https://doi.org/10.3390/molecules24122261
Molinari A, Oliva A, Arismendi-Macuer M, Guzmán L, Acevedo W, Aguayo D, Vinet R, San Feliciano A. Antiproliferative Benzoindazolequinones as Potential Cyclooxygenase-2 Inhibitors. Molecules. 2019; 24(12):2261. https://doi.org/10.3390/molecules24122261
Chicago/Turabian StyleMolinari, Aurora, Alfonso Oliva, Marlene Arismendi-Macuer, Leda Guzmán, Waldo Acevedo, Daniel Aguayo, Raúl Vinet, and Arturo San Feliciano. 2019. "Antiproliferative Benzoindazolequinones as Potential Cyclooxygenase-2 Inhibitors" Molecules 24, no. 12: 2261. https://doi.org/10.3390/molecules24122261