New Triterpenoid from Novel Triterpenoid 15-O-Glycosylation on Ganoderic Acid A by Intestinal Bacteria of Zebrafish
Abstract
:1. Introduction
2. Results
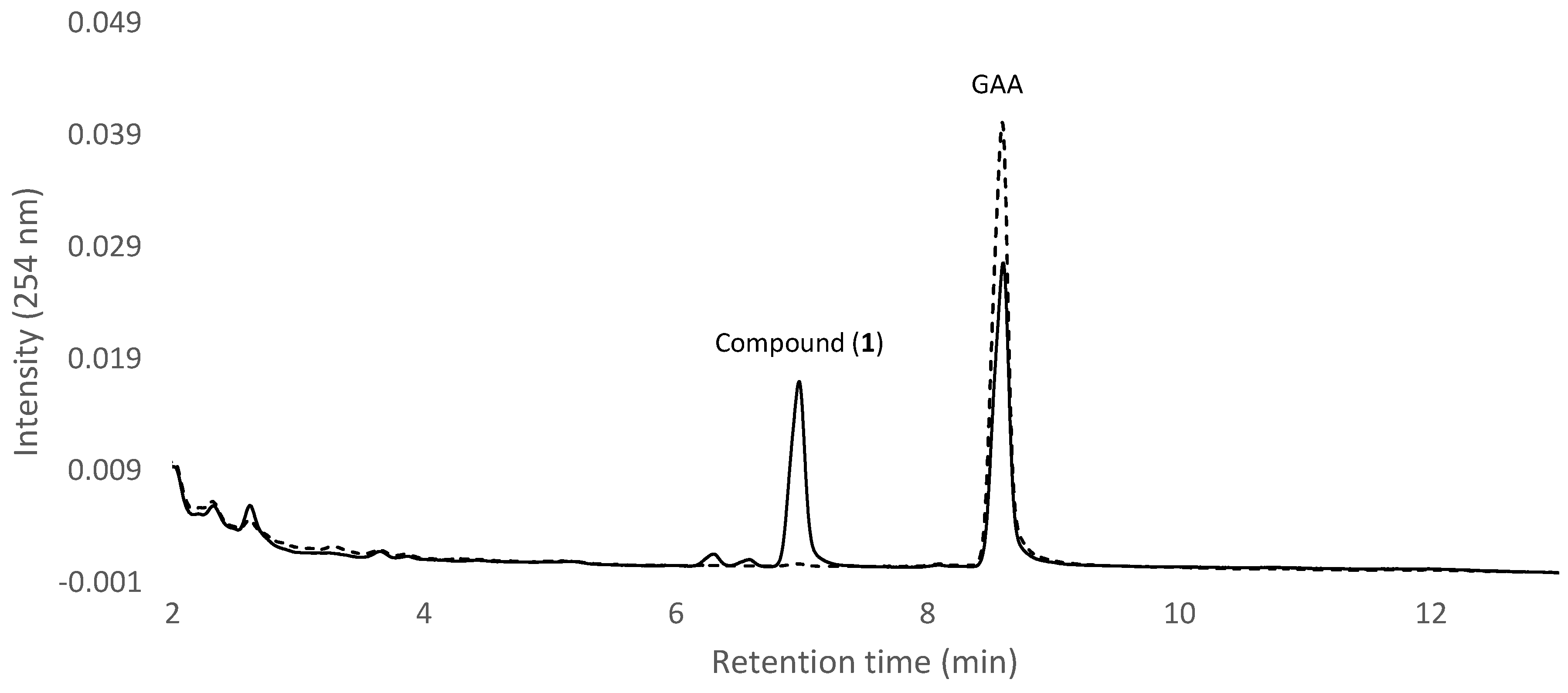
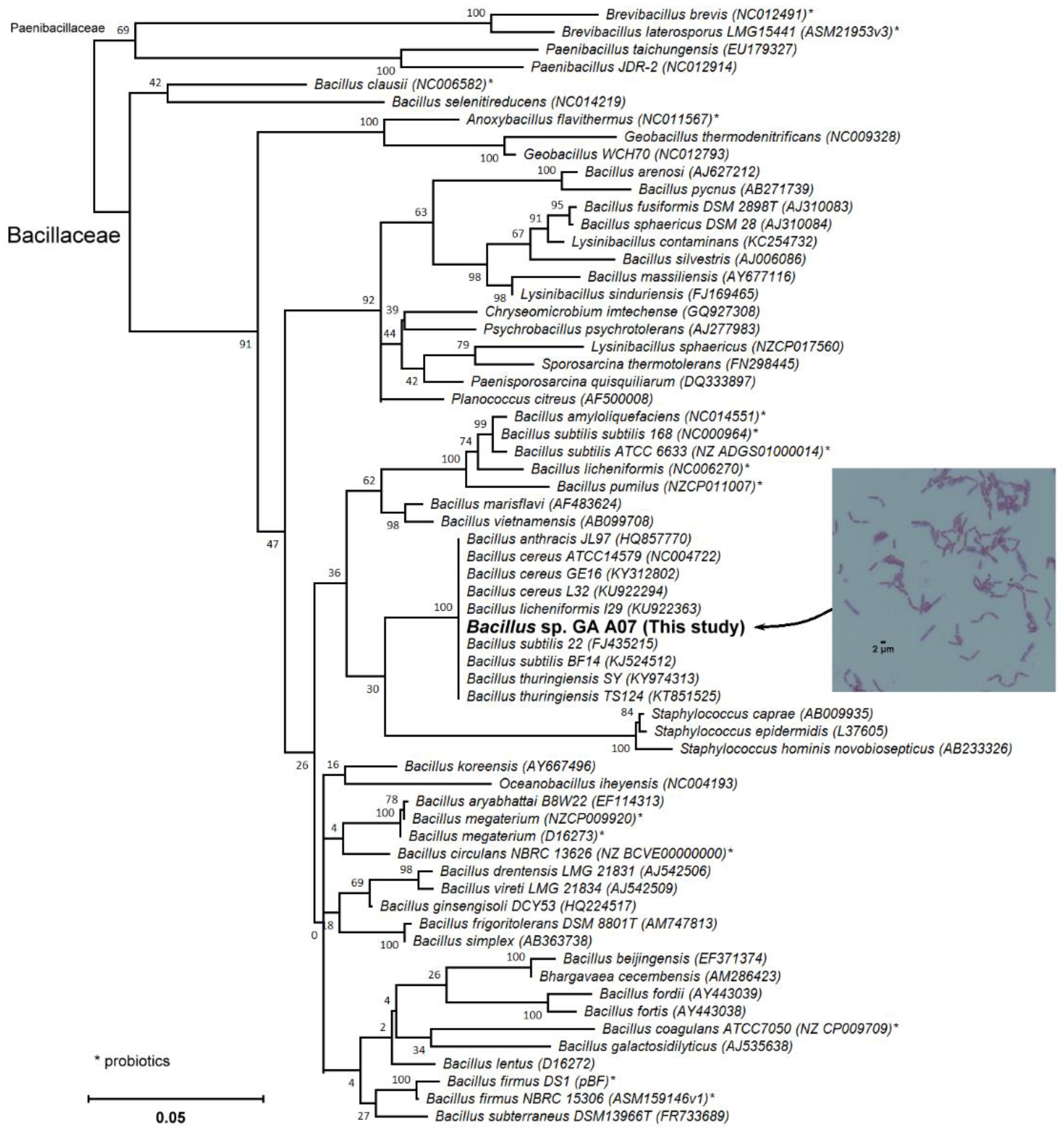
2.1. Screening and Identification of Intestinal Bacteria with Biotransformation Activity
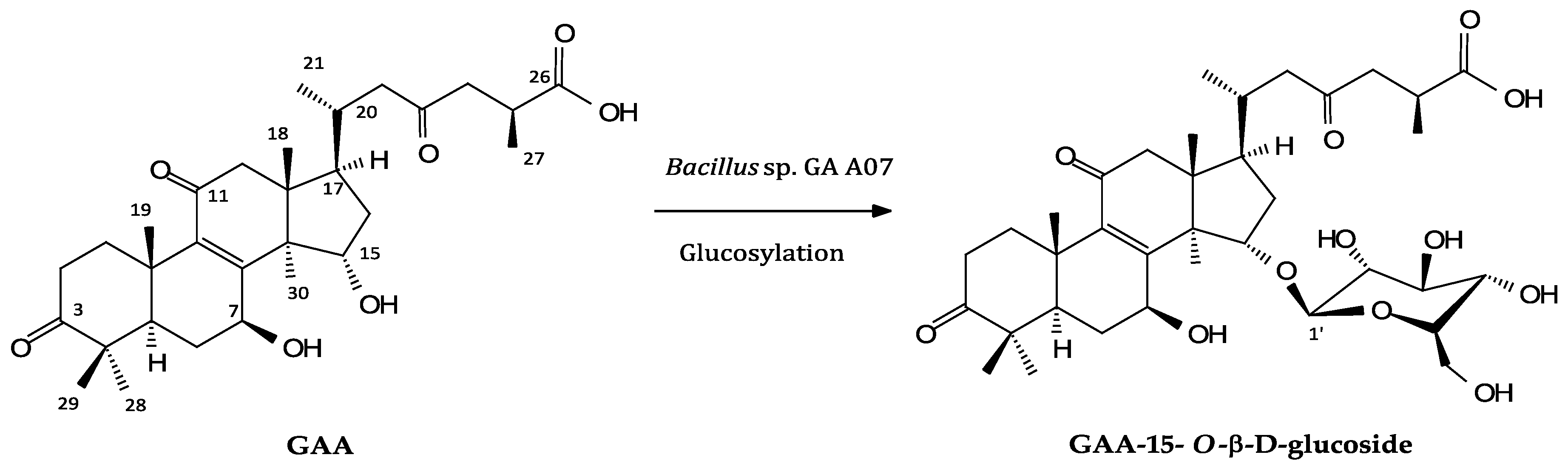
2.2. Isolation and Identification of Biotransformation Metabolite
3. Discussion
4. Materials and Methods
4.1. Microorganism and Chemicals
4.2. Screening and Identification of Intestinal Bacteria with Biotransformation Activity
4.3. UPLC Analysis
4.4. Candidate Strain Classification via 16S rRNA Gene Analysis
4.5. Confirming Possession of the pXO1 Plasmid
4.6. Scaled-Up Fermentation, Isolation, and Identification of the Biotransformation Products
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xu, J.W.; Zhao, W.; Zhong, J.J. Biotechnological production and application of ganoderic acids. Appl. Microbiol. Biotechnol. 2010, 87, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhang, H.; Sun, X.; Zhao, H.; Wu, L.; Zhu, D.; Yang, G.; Shao, Y.; Zhang, X.; Mao, X.; et al. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Asaka, Y.; Miura, I.; Mori, H. Structures of ganoderic acid A and B, two new lanostane type bitter triterpenes from Ganoderma lucidum (FR.) Karst. Helv. Chim. Acta 1982, 65, 611–619. [Google Scholar] [CrossRef]
- Jiang, J.; Grieb, B.; Thyagarajan, A.; Sliva, D. Ganoderic acids suppress growth and invasive behavior of breast cancer cells by modulating AP-1 and NF-κB signaling. Int. J. Mol. Med. 2008, 21, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Li, G.; Xu, H.; Lü, C. Inhibition of the JAK-STAT3 signaling pathway by ganoderic acid A enhances chemosensitivity of HepG2 cells to cisplatin. Planta Med. 2012, 78, 1740–1748. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, D.; Tai, J.; Wang, L. Ganoderic acid A inhibits proliferation and invasion, and promotes apoptosis in human hepatocellular carcinoma cells. Mol. Med. Rep. 2017, 16, 3894–3900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akihisa, T.; Nakamura, Y.; Tagata, M.; Tokuba, H.; Yasukawa, K.; Uchiyama, E.; Suzukli, T.; Kimura, Y. Anti-inflammatory and anti-tumor-promoting effects of triterpene acids and sterols from the fungus Ganoderma lucidum. Chem. Biodivers. 2007, 4, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Koyama, K.; Imaizumi, T.; Akiba, M.; Kinoshita, K.; Takahashi, K.; Suzuki, A.; Yano, S.; Horie, S.; Watanabe, K.; Naoi, Y. Antinociceptive components of Ganoderma lucidum. Planta Med. 1997, 63, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chang, Q.; Wong, L.K.; Chong, F.S.; Li, R.C. Triterpene antioxidants from Ganoderma lucidum. Phytother. Res. 1999, 13, 529–531. [Google Scholar] [CrossRef]
- Sultana, N.; Saify, Z.S. Enzymatic biotransformation of terpenes as bioactive agents. J. Enzym. Inhib. Med. Chem. 2013, 28, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Muffler, K.; Leipold, D.; Scheller, M.C.; Haas, C.; Steingroewer, J.; Bley, T.; Neuhaus, H.E.; Mirata, M.A.; Schrader, J.; Ulber, R. Biotransformation of triterpenes. Process Biochem. 2011, 46, 1–15. [Google Scholar] [CrossRef]
- Parra, A.; Rivas, F.; Garcia-Granados, A.; Martinez, A. Microbial transformation of triterpenoids. Mini-Rev. Org. Chem. 2009, 6, 307–320. [Google Scholar] [CrossRef]
- Shah, S.A.A.; Tan, H.L.; Sultan, S.; Faridz, M.A.B.M.; Shah, M.A.B.M.; Nurfazilah, S.; Hussain, M. Microbial-catalyzed biotransformation of multifunctional triterpenoids derived from phytonutrients. Int. J. Mol. Sci. 2014, 15, 12027–12060. [Google Scholar] [CrossRef] [PubMed]
- Cahill, M.M. Bacterial-flora of fishes: A review. Microb. Ecol. 1990, 19, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Huber, I.; Spanggaard, B.; Appel, K.F.; Rossen, L.; Nielsen, T.; Gram, L. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum). J. Appl. Microbiol. 2004, 96, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Lescak, E.A.; Milligan-Myhre, K.C. Teleosts as model organisms to understand host-microbe interactions. J. Bacteriol. 2017, 199, e00868-16. [Google Scholar] [CrossRef] [PubMed]
- Reinoso Webb, C.; Koboziev, I.; Furr, K.L.; Grisham, M.B. Protective and pro-inflammatory roles of intestinal bacteria. Pathophysiology 2016, 23, 67–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wu, Y.; Wang, Y.Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant properties of probiotic bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Zorriehzahra, M.J.; Delshad, S.T.; Adel, M.; Tiwari, R.; Karthik, K.; Dhama, K.; Lazado, C.C. Probiotics as beneficial microbes in aquaculture: An update on their multiple modes of action: A review. Vet. Q. 2016, 36, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Chandranaik, B.M.; Giridhar, P.; Muniyellappa, H.K.; Hegde, R.; Earanna, N.; Rathnamma, D.; Kalge, R.S.; Kanaka, S.; Chandrakala, G.K.; Ashamayanna; et al. Bacillus cereus harboring the pXO1 palsmid with pag gene causes anthrax-like fatal septicemia in immunosuppressed cattle. Vet. Arh. 2015, 85, 347–357. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; ISBN 0195135857. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kohda, H.; Tokumoto, W.; Sakamoto, K.; Fujii, M.; Hirai, Y.; Yamasaki, K.; Komoda, Y.; Nakamura, H.; Ishihara, S. The biologically active constituents of Ganoderma lucidum (FR.) Karst. histamine release-inhibitory triterpenes. Chem. Pharm. Bull. 1985, 33, 1367–1374. [Google Scholar] [CrossRef]
- Chang, T.S.; Chiang, C.M.; Siao, Y.Y.; Wu, J.Y. Sequential biotransformation of antcin K by Bacillus subtilis ATCC 6633. Catalysts 2018, 8, 349. [Google Scholar] [CrossRef]
- Wang, W.W.; Xu, S.H.; Zhao, Y.Z.; Zhang, C.; Zhang, Y.Y.; Yu, B.Y.; Zhang, J. Microbial hydroxylation and glycosylation of pentacyclic triterpenes as inhibitors on tissue factor procoagulant activity. Bioorg. Med. Chem. Lett. 2017, 27, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, S.H.; Ma, B.L.; Wang, W.W.; Yu, B.Y.; Zhang, J. New derivatives of ursolic acid through the biotransformation by Bacillus megaterium CGMCC 1.1741 as inhibitors on nitric oxide production. Bioorg. Med. Chem. Lett. 2017, 27, 2575–2578. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zou, Z.; Fu, S.; Sun, L.; Su, Z.; Sun, D. Microbial oxidation and glucosidation of echinocystic acid by Nocardia coralline. J. Mol. Catal. B Enzym. 2010, 66, 219–223. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Yang, J.; Zhu, Y.; Men, Y.; Zeng, Y.; Cai, Y.; Dong, C.; Dai, Z.; Zhang, X.; et al. Use of a promiscuous glycosyltransferase from Bacillus subtilis 168 for the enzymatic synthesis of novel protopanaxtriol-type ginsenosides. J. Agric. Food Chem. 2017, 66, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Li, J.; Yao, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Yang, J.; Sun, Y. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J. Biotechnol. 2017, 248, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Hu, Z.; Zhang, T.; Gong, T.; Chen, J.; Zhu, P.; Li, Y.; Yang, J. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab. Eng. 2017, 44, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Jin, Y.; Wang, C.; Kim, Y.J.; Perez, Z.E.J.; Baek, N.I.; Mathiyalagan, R.; Markus, J.; Yang, D.C. Rare ginsenoside Ia synthesized from F1 by cloning and overexpression of the UDP-glycosyltransferase gene from Bacillus subtilis: synthesis, characterization, and in vitro melanogenesis inhibition activity in BL6B16 cells. J. Gingeng Res. 2018, 42, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.M.; Abd-El-Rahman, A.M.; John, G.; Mohamed, M.F. Characterization of some bacteria isolated from Oreochromis niloticus and their potential use as probiotics. Aquaculture 2008, 277, 1–6. [Google Scholar] [CrossRef]
- Avella, M.A.; Gioacchini, G.; Decamp, O.; Makridis, P.; Bracciatelli, C.; Carnevali, O. Application of multi-species of Bacillus in sea bream larviculture. Aquaculture 2010, 305, 12–19. [Google Scholar] [CrossRef]
- He, S.X.; Zhang, Y.; Xu, L.; Yang, Y.L.; Marubashi, T.; Zhou, Z.G.; Yao, B. Effects of dietary Bacillus subtilis C-3102 on the production, intestinal cytokine expression and autochthonous bacteria of hybrid tilapia Oreochromis niloticus female × Oreochromis aureus male. Aquaculture 2013, 412, 125–130. [Google Scholar] [CrossRef]
- Sorokulova, I.B.; Pinchuk, I.V.; Denayrolles, M.; Osipova, I.G.; Huang, J.M.; Cutting, S.M.; Urdaci, M.C. The safety of two Bacillus probiotic strains for human use. Dig. Dis. Sci. 2008, 53, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Quan, K.; Liu, Q.; Wan, J.Y.; Zhao, Y.J.; Guo, R.Z.; Alolga, R.N.; Li, P.; Qi, L.W. Rapid preparation of rare ginsenosides by acid transformation and their structure-activity relationships against cancer cells. Sci. Rep. 2015, 5, 8598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.M.; Wang, T.Y.; Ke, A.N.; Chang, T.S.; Wu, J.Y. Biotransformation of ergostane triterpenoid antcin K form Antrodia cinnamomea by soil-isolated Psychrobacillus sp. AK 1817. Catalysts 2017, 7, 299. [Google Scholar] [CrossRef]
Sample Availability: Sample of the compound GAA-15-O-β-glucoside is available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, T.-S.; Chiang, C.-M.; Wang, T.-Y.; Lee, C.-H.; Lee, Y.-W.; Wu, J.-Y. New Triterpenoid from Novel Triterpenoid 15-O-Glycosylation on Ganoderic Acid A by Intestinal Bacteria of Zebrafish. Molecules 2018, 23, 2345. https://doi.org/10.3390/molecules23092345
Chang T-S, Chiang C-M, Wang T-Y, Lee C-H, Lee Y-W, Wu J-Y. New Triterpenoid from Novel Triterpenoid 15-O-Glycosylation on Ganoderic Acid A by Intestinal Bacteria of Zebrafish. Molecules. 2018; 23(9):2345. https://doi.org/10.3390/molecules23092345
Chicago/Turabian StyleChang, Te-Sheng, Chien-Min Chiang, Tzi-Yuan Wang, Chun-Hsien Lee, Yu-Wen Lee, and Jiumn-Yih Wu. 2018. "New Triterpenoid from Novel Triterpenoid 15-O-Glycosylation on Ganoderic Acid A by Intestinal Bacteria of Zebrafish" Molecules 23, no. 9: 2345. https://doi.org/10.3390/molecules23092345
APA StyleChang, T.-S., Chiang, C.-M., Wang, T.-Y., Lee, C.-H., Lee, Y.-W., & Wu, J.-Y. (2018). New Triterpenoid from Novel Triterpenoid 15-O-Glycosylation on Ganoderic Acid A by Intestinal Bacteria of Zebrafish. Molecules, 23(9), 2345. https://doi.org/10.3390/molecules23092345







