Abstract
Lactobacillus plantarum is one of the most predominant species in the human gut microbiota of healthy individuals. We have previously characterized some probiotic features of L. plantarum LM3, as the high resistance to different stress, the binding ability toward some extracellular matrix proteins and plasminogen and the immunomodulatory role of the surface expressed adhesin EnoA1. We have also identified the flmA, flmB and flmC genes, coding for putative proteins named FlmA, FlmB and FlmC, whose null mutations partially impaired biofilm development; the L. plantarum LM3–6 strain, carrying a deletion in flmC, showed a high rate of autolysis, supporting the hypothesis that FlmC might be involved in cell wall integrity. Here, we report the in-silico characterization of ΔTM-FlmC, a portion of the FlmC protein. The protein has been also expressed, purified and characterized by means of CD spectroscopy, ICP-mass and UHPLC-HRMS. The obtained experimental data validated the predicted model unveiling also the presence of a bound lipid molecule and of a Mg(II) ion. Overall, we provide strong evidences that ΔTM-FlmC belongs to the LytR-CpsA-Psr (LCP) family of domains and is involved in cell envelope biogenesis.
1. Introduction
Studies on biofilm development are of great interest for the impact they may have on different aspects of the human life [1,2,3,4]. Exploitation of microbial biofilm has been accomplished in different fields, including bioremediation and biotechnological production processes [3,5,6]. Contrariwise, biofilms developed by pathogens represent a serious problem for human health; indeed, cells resident into biofilms show an increased resistance to environmental stress, to immune-response and to antimicrobial molecules [7]. In a continuous struggle to discover new antibiotics or new formulations [8,9,10,11], the scientific community has been making big efforts to search for molecules capable to treat biofilm-depending chronic infections [12,13]. On the other hand, commensals belonging to the microbiota of healthy individuals may grow in the sessile form associated to the mucus layer in the gut and to particulate surfaces in the gut lumen, acting as innate immune protectors of the underlying epithelial cells and as antagonists of pathogens by means of competitive exclusion mechanisms [14,15]. Moreover, Lactobacillus biofilms growing on vaginal or intestinal epithelia may have a protecting role against sexually transmitted, urinary or intestinal infections in healthy individuals [15,16,17]. Therefore, one of the features searched in probiotic strains to be selected for treatment of vaginal or intestinal dysbiosis is the ability to form biofilms in the environmental conditions encountered either in the vagina or in the colon [4,18]. Indeed, it is well known that the physico-chemical conditions have significant influences on the ability of the different strains to form biofilms [17].
Probiotics, whose features are defined on the strain basis, mainly belong to the group of the lactic acid bacteria of the genus Lactobacillus and Bifidobacterium. Among these, some strains of Lactobacillus plantarum, one of the most predominant species in the human gut microbiota of healthy individuals, have been defined as good performing probiotic microorganisms [19,20,21,22]. L. plantarum is able to colonize a wide range of environmental niches for its high metabolic versatility, contrariwise to the majority of probiotic lactobacilli, which are highly specialized for growth in a limited number of conditions [23]. Comparative genomics of various L. plantarum strains revealed high genomic diversity, based on the presence of genomic islands containing mosaic modules of genes for carbohydrate utilization, shedding light on the capability of various L. plantarum strains to colonize different environmental niches [24]. Based on these features, L. plantarum is largely used as starter in food industry for vegetable, meat, fodder and milk fermentation and for the development of probiotic formulations [24,25].
We have previously characterized important probiotic features of L. plantarum LM3, as the high resistance to different stress, assessed in a multiple test for a simulated gastrointestinal transit (Vastano, personal communication), the binding ability toward some extracellular matrix proteins and plasminogen and the immunomodulatory role of a surface expressed adhesin, namely the EnoA1 protein, also involved in biofilm development [26,27,28,29]. Moreover, we have identified the flmA, flmB and flmC genes, coding for putative proteins named FlmA, FlmB and FlmC, whose amino acid sequences show significant percentage of identity with Streptococcus mutans BrpA (biofilm regulator protein A) [30,31]. BrpA is located on the cell surface and is involved in maintaining the structure of the cell wall through the regulation of autolysins. More recently BrpA has been indicated as a virulence factor in different pathogenic streptococcal species [32,33]. Indeed, L. plantarum strains carrying null mutations in the flmA, flmB and flmC genes, were partially impaired in biofilm development; one of them, the L. plantarum LM3–6 strain, carrying a deletion in flmC, showed a high rate of autolysis, supporting the hypothesis that FlmC might be involved in cell wall integrity [30].
Interestingly, FlmA, FlmB and FlmC, as well as BrpA, contain a highly conserved domain, that appears to belong to the LytR-CpsA-Psr (LCP) family of domains. The LCP family of cell envelope-associated transcriptional attenuators gained attention upon the discovery that some members of this family influence various virulence factors as well as antibiotic resistance of important human pathogens. Moreover, the LCP family seems to play a role in bacterial cell envelope maintenance [31,34,35,36]. Members of the LCP family were demonstrated to be magnesium-dependent phosphotransferase, responsible of the linkage of the anion cell-wall attached polymers to the peptidoglycan [37]. In this framework, the characterization of FlmC, especially if integrated with studies on its structural features, appears to be of interest, being L. plantarum a protective probiotic as quoted above.
In this study, we report the expression, purification and characterization of the L. plantarum ΔTM-FlmC, a portion of the FlmC protein. In order to describe the structural characteristics of this protein, we first performed an in-silico analysis and a computational modelling study. The protein has been then characterized by means of CD spectroscopy, ICP-mass and UHPLC-HRMS. The experimental data obtained validated the predicted model unveiling also the presence of a bound lipid molecule and of a Mg(II) ion. Overall, we provide strong evidences that ΔTM-FlmC belongs to the LCP family and is involved in cell envelope biogenesis.
2. Results and Discussion
2.1. Sequence Analysis and ΔTM-FlmC Expression
Recently, we have identified the flmA, flmB and flmC genes from L. plantarum LM3, coding for putative proteins named FlmA, FlmB and FlmC [30]. The secretome database attributes to these ORF a role of regulatory proteins and a function of cell envelope-related transcriptional attenuator; it is also known that all of them contain a N-terminal trans-membrane anchor domain. Interestingly, the three L. plantarum proteins contain in their C-terminal portion highly conserved regions that closely resemble the family of the LytR-CpsA-Psr (LCP) domain. Specifically, among the three Flm proteins, we have chosen to characterize FlmC, as the deletion of the corresponding gene (flmC) results in a L. plantarum strain (L. plantarum LM3–6) with a compromised biofilm development and increased autolytic activity phenotypes; the deletion of the other flm genes only hampered biofilm development but not the autolytic activity [30].
Figure 1 proposes the primary sequence of FlmC and its alignment, using the server ClustalO, with the LCP domain of the protein Cps2A. The server aligns the two proteins providing 50% of similarity and 29% of identity between the two sequences, especially in the region starting from residue V81 of FlmC and residue L219 of Cps2A.

Figure 1.
The primary sequence of L. plantarum FlmC aligned with the protein Cps2A from S. pneumoniae.
This alignment underlines the high similarity of the two proteins in the portion that contains the LCP domain.
The X-ray structure of the protein ΔTM-Cps2A from Streptococcus pneumonia, obtained by deletion of the transmembrane portion of Cps2A and containing the LCP domain and an accessory domain, was published in 2011 [37] shedding light on the structural details and function of this domain.
We thus decided to further characterize the L. plantarum FlmC by expressing and purifying a portion of the protein. One of the requisite to attend in selecting the appropriate portion of the protein to express is to avoid inclusion of low-complexity regions or hydrophobic residue rich sequences at the C- or N-termini [38]. For this reason, we have studied the aggregation profile of the selected protein using the servers AGGRESCAN [39] and TMHMM [40], whose results are reported in Figure 2.
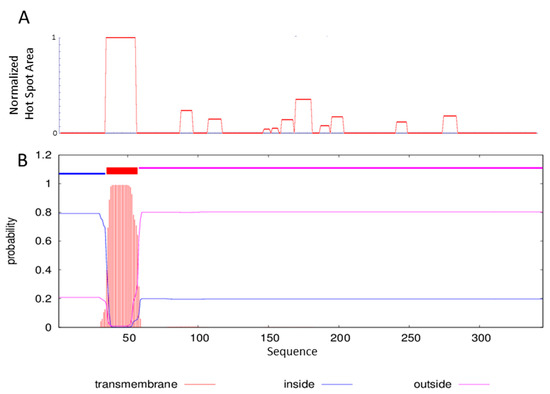
Figure 2.
FlmC aggregation profile using the servers AGGRESCAN (A). Prediction of transmembrane helices by TMHMM server (B).
The region spanning residues 33–56 is clearly characterized by a highly aggregation prone sequence (Figure 2-panel A). The analysis performed with the server TMHMM capable of predicting the formation of transmembrane helices in proteins outlines for the same region the possibility of a transmembrane domain (Figure 2-panel B).
Therefore, we expressed and purified the sequence spanning residues 81 to 335 obtaining the deletion mutant FlmC81-335 (ΔTM-FlmC). For high-level protein production purposes, we expressed the protein in E. coli BL21(DE3) that has the advantage of being deficient in both lon and ompT genes coding for proteases and it is compatible with the T7 lacO promoter system. For purification purposes, the protein was produced with a hexahistidine tag that was proven not to consistently impact on the terminal structure of recombinant proteins.
2.2. ΔTM-FlmC Structural Model
The three-dimensional structure represents a very informative and useful tool to understand the functional features of the examined proteins. ΔTM-FlmC three-dimensional structure (Figure 3) was modelled using the I-TASSER algorithm [41] with the primary sequence as input data, using as a template a member of the LCP protein family (3TFL pdb code), taking advantage of a 29% sequence identity for the full-length sequence alignment among the two systems (up to 37% identity at the β-sheet level). In particular, the structure prediction by I-TASSER relies on template known protein structures and the prediction procedure is based on matching the query sequence against a non-redundant sequence database. The computational model of ΔTM-FlmC gave a C-scores of 1.53 indicating a good quality of the predicted model. The c-score that estimates the quality of I-TASSER predicted models typically ranges between −5 and 2, with higher value indicating models with a higher confidence.
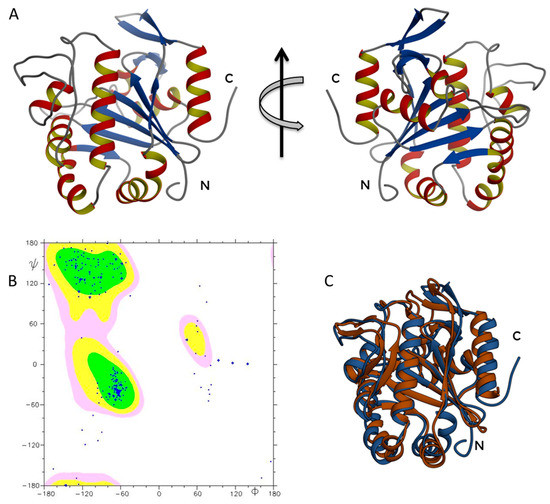
Figure 3.
(A) ΔTM-FlmC structural model; (B) the Ramachandran plot confirms the good quality of ΔTM-FlmC predicted model; (C) ΔTM-FlmC superimposition with the crystallographic structure of Cps2A.
The obtained ΔTM-FlmC structure (Figure 3A) was further analysed by evaluating the Ramachandran plot (Figure 3B), using the software PROCHECK e MOLMOL. Over 97% of the residues were either in favoured or allowed regions denoting the good quality of the predicted model. The ΔTM-FlmC model was then energetically minimized by using Gromacs [42].
In order to gain further insight into the stability of the modelled structure we ran 5 ns MD simulation as reported in the methods section. The last frame of the trajectory was used for further analyses.
As expected, ΔTM-FlmC adopted a compact structure similar to the LCP domain so far characterized (Figure 3C). Overall, ΔTM-FlmC exhibits the typical topology of this domain with most of the hydrophobic amino acid residues buried in the interior and many of the polar residues on the surface. Hydrophobic interactions are a major force that drive protein folding and structure by forcing hydrophobic side chains to closely associate in such a way that they result shielded from polar solvents [43,44,45]. The tertiary structure of ΔTM-FlmC had an architecture with a central sheet composed by six-strands surrounded on both faces by ten (total) -helices held together by short non-helical regions (Figure 3A).
The overall architecture provided a structural support for a hydrophobic pocket (Figure 3A,B) between the main-sheet and helices 3–7 in which, in the Cps2A protein [37], a polyisoprenoid phosphate lipid is inserted.
A comparison of ΔTM-FlmC predicted model with the crystallographic structure of Cps2A (PDB code: 2XXP—Figure 3C) indicates that both proteins show a classical globular fold. The back-bone superposition of ΔTM-FlmC model with the portion encompassing residues 219–481 of the crystallographic structure showed an RMSD of 2.1 Å. Predictable minor differences were found between the two protein structures, being local rearrangements the response to preserve the global fold in presence of differences in amino acids composition [46,47]. Therefore, we compared the orientation and length of the secondary structure elements in the modelled structure with those of the reported structure of the LCP domain, respectively. As reported in Figure 4, in ΔTM-FlmC α-helix 6 and 3–10 helix 7 (L178-D182 and F183-Q190, respectively-panel A), α-helix 8 and α-helix 9 (F197-T205 and R208-T211-panel B) and α-helix 10 (S237-S249-panel C) slightly differ in terms of length and axis orientation with respect to the comparable secondary structural elements of Cps2A. Notably, the two proteins share a similar hydrophobic cleft, in terms of structural features. Overall, the hydrophobic pocket in which is located the polyisoprenoid phosphate lipid, considering also the sidechain orientation of the distal and proximal histidine residues (H64, H93), does not show any significant structural difference.
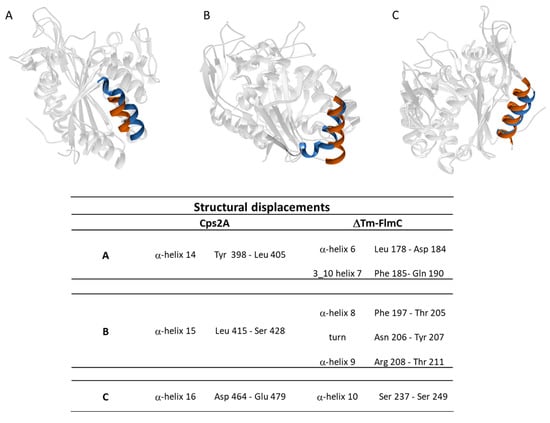
Figure 4.
Panel A, B and C show the main structural differences between ΔTM-FlmC (blue) and Cps2A (red); the structural elements shown are reported in the table.
2.3. ΔTM-Flmc Binding Pocket
Predictions of the binding pocket and docking studies were carried out on the final protein conformation of the MD simulation (rmsd = 0.4 nm on all atoms along the 5 ns simulation time). A combinatorial computational strategy was used in order to better validate our modelled system.
First, the identification of pockets on protein surface was performed by LIGSITE [48] and 3 dligandsite [49], independently and prior to docking studies. The two chosen predictors rely on different search algorithms, the former based on solvent accessibility analysis by Connolly surface approach, the latter predicting ligand binding sites based on ligands present in homologous protein structures. The combination of the different outcomes yielded to the definition of a protein cavity running along the protein, involving both charged amino acids from the top of the β-sheet (D29, R145, R147, R157) and hydrophobic residues buried at the α-helix level (namely I165, I168, I169, L187, V204). It is interesting that very bulky and hydrophobic amino acids localized at the inner helical region of the identified pocket result highly aligned/overlapping to residues that make contacts with prenyl moiety onto homologous Cps2A complex.
To further support these findings (and provide a visual idea to the model), we investigated putative binding poses by means of docking studies (Figure 5). Octaprenyl pyrophosphate ligand (from 3TFL PDB) was docked onto the protein, with no constraints on the binding pocket using SwissDock [50]. More than 250 binding modes of the ligand into ΔTM-FlmC cavity were generated. From Figure 5 and Figure 6, it is evident that phosphate heads in all poses mostly overlap at the top of the β-sheet interacting with D29, R145, in a similar rearrangement to what was observed for Cps2A homologous protein.
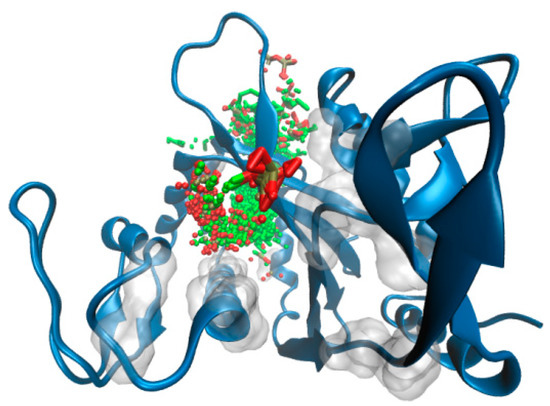
Figure 5.
SwissDock binding predictions. The full set of octaprenyl pyrophosphate docking poses are shown in sticks at the binding pocket. Transparent surface is used to pinpoint residues that define the binding cavity. ΔTM-FlmC is rendered in blue cartoons.
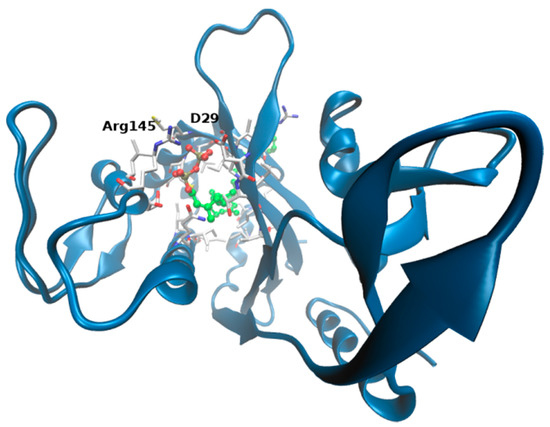
Figure 6.
Docking models. Best ranked ligand pose predicted by SwissDock: ΔTM-FlmC is shown in blue cartoons, octaprenyl pyrophosphate in CPK and amino acids within 3 Å from the ligand in white sticks. Interacting residues at the top of the cavity are labelled (D29, R145). See main text.
Presumably, on the other side, long aliphatic chain inserts into the channel delimited by the β-sheet and the long α-helix, making contacts with I165, L187 localized at the very buried cavity. The best ranked binding pose predicted is displayed in Figure 6 (the docking pose is associated to a free energy of binding of −12.7 kcal/mol).
An additional validation of the bound complex was also taken from GalaxyWEB [51]. This server is able to provide predictions of ligands that are likely to bind to the protein and their relative binding poses. Figure S1 shows binding site interactions analysis predicted by Galaxy on the first docking pose. It is interesting to underline that the main interactions made by the phosphate moieties at the top of the binding site are conserved between the two complexes.
2.4. Circular Dichroism Characterization and Validation of the Obtained Model
One strategy for assessing accuracy is to cross-validate the calculated structures or models using properties not considered in the computation. We performed a cross-validation analysis for ΔTM-FlmC predicted models using the Circular Dichroism (CD) to further validate the predicted model. CD is an excellent tool for rapid determination of secondary structure and folding properties of synthesized peptides [8,11,52,53] and of proteins [54,55] that have been obtained using recombinant techniques or purified from tissues. The most widely used applications of protein CD are to determine whether an expressed, purified protein is folded, or if a mutation affects its conformation or stability [47]. In addition, it can be used to study protein and peptide in teractions [56]. The CD spectrum of ΔTM-FlmC (Figure 7A) is characteristic of a well-structured protein containing both -helical and -sheet secondary structure. We estimated using the CD data the protein secondary structure content by using the server BeStSel (Figure 7B). The data indicate that the secondary structure content of our purified protein well fits the content of secondary structure evaluated for the I-TASSER calculated model. In fact, purified ΔTM-FlmC-helix content calculated from the CD spectra is 30.3% while the structures content is 26.7%. These values are in a good agreement with the -helix and -structures amounts obtained evaluating the predicted model using the software MOLMOL and DSSP.
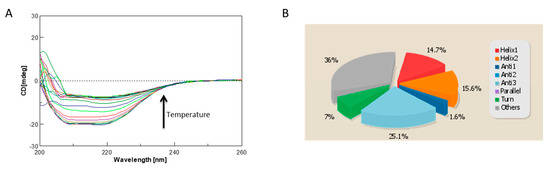
Figure 7.
(A) CD spectra of ΔTM-FlmC reported as function of temperature; (B) Secondary structure content evaluated using the CD data by the server BeStSel.
Overall, our analysis demonstrated that the I-TASSER computationally obtained model represents a realistic picture of the tertiary structure that the purified ΔTM-FlmC protein adopts in solution. The stability of ΔTM-FlmC has been also evaluated by monitoring the CD signal at 222 nm (Figure 7A). ΔTM-FlmC irreversible thermal unfolding encompassing the temperature range between 278 and 353 K gave a midpoint temperature of unfolding at around 315 K confirming an overall structural stability of the purified protein.
2.5. ΔTM-FlmC Domain Binds a Lipid Molecule and a Magnesium Ion
Kaway et al. [37] showed that the Cps2A protein co-purifies with a polyisoprenoid phosphate lipid located in a hydrophobic pocket between the main -sheet and -helices 3–7. They hypothesized that their protein had bound the lipid when heterologously expressed in E. coli, confirming its affinity for a lipid-linked capsule precursor in S. pneumoniae. The lipid was built as a monotrans, octa-cis decaprenyl-phosphate. For the ΔTM-FlmC protein, the server I-TASSER, able to foretell binding sites in the calculated models, predicted an extensive hydrophobic binding pocket that could easily accommodate an analogous lipid molecule.
Figure 6 shows ΔTM-FlmC cavity: it is composed by hydrophobic side chains (Figure S1) of residues that, while not completely identical to those of Cps2A, are conserved across the LCP family and are likely to give to the pocket the same hydrophobic character. Moreover, ΔTM-FlmC cavity appears to have a comparable diameter so that a lipid molecule can be easily accommodated in it with only few interactions with the protein (Figure 6). Figure S1 shows the protein residues that constitute the hydrophobic pocket and are likely to be involved in the interaction with the lipid molecule.
As for Cps2A, also in the case of ΔTM-FlmC charged residues (R50, R145 and R157) surround the area where is located the phosphate head group of the prenyl, stabilizing the binding.
Given these evidences, we have investigated whether also ΔTM-FlmC co-purified with a similar lipid. We performed an organic solvent extraction as in Harrison et al. [57] and evaluated the products via TLC (thin-layer chromatography) analysis that suggested the presence of a lipid (data not shown). The total ion current (TIC) chromatogram (Figure S2A), acquired in positive ion mode in the range m/z 100–1500, showed a protonated molecular ion at m/z 775.5789. Based on previous findings [1], the occurrence of a decaprenyl phosphate (dpr-P) ligand tightly bound to the protein was hypothesized. Indeed, the [M + H]+ ion was in accordance with an in-source formed dehydro dpr-P (Figure S2A), whose proposed structure was as depicted. The detection of the doubly charged ion at m/z 387.2863 ([M−2H]2−), in accordance with the molecular formula C50H81O4P2−, recorded in the TOF-MS spectrum acquired in negative ion mode, likely confirmed dpr-P presence (Figure S2B).
Members of the LCP family were proved to bind a magnesium ion which appears to be necessary for their magnesium-dependent phosphotransferase activity.
As proposed by Kawai and co-workers [37] a Mg(II) ion binds the Cps2A protein with an octahedral geometry. The protein interacts with the metal ion using the two aspartate residues D234 and D246. The magnesium ion plays a crucial role in the function of the protein as its loss, as well as the mutation of one of the coordinating aspartates, leads to a reduction in the phosphatase activity. ΔTM-FlmC three-dimensional structural model evidences how the two homologous positions are occupied also in this protein by two aspartates (D16 and D29), suggesting for these residues the same role in coordinating a magnesium ion. We have thus explored the presence of metal ions bound to ΔTM-FlmC by ICP-MS that revealed how also this protein binds a Mg(II) ion.
Our experiments conducted on different dilutions of the protein solution indicated the presence of magnesium ions whose concentrations varied as function of the protein concentrations (Figure S3), thus reinforcing the idea that D16 and D29 are likely to be involved in the coordination of a Mg(II) ion.
3. Materials and Methods
3.1. Bioinformatics
All the used amino acid sequences were retrieved and analysed using the BLAST software (http:// blast.ncbi.nlm.nih.gov/Blast.cgi). Alignments were performed by Clustal Omega at EMBnet-CH (http://www.ebi.ac.uk/Tools/msa/clustalo). FlmC aggregation profile was evaluated using the server AGGRESCAN and transmembrane helix content evaluated by the server TMHMM [40]. ΔTM-FlmC three-dimensional structure has been modelled by I-Tasser [41]. Molecular dynamics simulations were run using Gromacs package (v.4.5.5) [58]. The secondary structure content from the CD data were evaluated using BestSel [59]. The structure has been validated using Procheck [60] and visualized using Pymol [61] and Chimera [62].
3.2. Expression and Purification of ΔTM-FlmC from L. plantarum
L. plantarum LM3 chromosomal DNA was used as template in a PCR reaction and a fragment of the flmC gene from nucleotides 241 to 1006 was amplified. Due to the presence of the NdeI restriction site in the flmC gene, a two steps-cloning strategy was used. The MLF1for/MLF2 rev and MLF3 for/MLF4 rev pairs of primers were used in the first and in the second step, respectively. The two DNA fragments of 402 and 363 bp, coding for aa residues V81–M215 and M215–A335, respectively, were amplified by PCR and cloned in pET22b(+) (Novagen). The 402 bp fragment was cloned in NdeI/NotI sites, yielding the pFlmC134 plasmid. The 363 bp fragment was cloned in the NdeI site of the recombinant pFlmC134, yielding the pFlmC254 plasmid, which was selected in E. coli TOP10. After sequencing, the recombinant pFlmC254 plasmid was used to transform E. coli BL21 (DE3) for FlmC81–335 (hereafter ΔTM-FlmC) expression. High protein yield was obtained by growing cells in 100 mL of liquid LB at 37 °C up to OD600 = 0.2. Cells were then centrifuged at 4.000× g for ten minutes and the pellet was re-suspended in 1L M9 medium and grown at 37 °C with shaking up to OD600 = 0.6, before being induced with 1 mM IPTG and incubated at 16 °C for 2 h. Cells were harvested by centrifugation at 4000× g for 30 min and washed with 50 mL of 20 mM Tris-HCl, pH 8.0, before a final centrifugation step at 4000× g for ten minutes. The cell pellet was then re-suspended in 50 mL buffer A (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM imidazole) before sonication for 5 min on ice by a 15 s on, 15 s off cycle. The lysed cells were clarified by centrifugation at 20,000× g for 30 min. The cell free extract was loaded onto a 3 mL Ni-charged resin (Biorad), for His-tagged affinity chromatography. The protein was eluted with 10 mL of buffer B (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 500 mM imidazole). The presence of ΔTM-FlmC was monitored by absorbance at 280 nm and confirmed by analysis of apparent molecular weight of the eluted protein by SDS-PAGE. UV-vis spectroscopy measurements were conducted using a UV-1700 Spectrometer (Shimadzu, Japan) [9] with 1 cm matched quartz cuvettes at room temperature in the wavelength range 200–500 nm.
3.3. Circular Dichroism
Circular Dichroism (CD) analysis was performed using a JASCO-815 CD (Jasco, USA) spectropolarimeter equipped with Peltier temperature control. ΔTM-FlmC samples were prepared in 20 mM Tris, 200 mM NaCl, pH 8.0. The data were collected using a quartz cuvette with a 1 cm path-length in the 190–260 nm wavelength range, a bandwidth of 1 nm, scanning speed of 50 nm min−1 and normalized against reference spectra to remove the buffer background contribution. The CD spectra have been de–convoluted by using the server BeStSel [59].
Thermal unfolding was followed recording CD spectra measured at 5 K intervals in the 278–353 K range. After the final measurement at 353 K, the sample was cooled back to 298 K and a final spectrum recorded.
3.4. 3D Structural Models
The 3D models for ΔTM-FlmC were obtained on the basis of its amino acid sequence using the I-TASSER software. I-TASSER (Interactive Threading ASSEmbly Refinement) is a computational method that combines all three conventional methods for structure modelling: comparative modelling, threading and ab initio modelling [41]. The c-score is a confidence value used to evaluate the quality of I-TASSER predicted models. It is estimated on the basis of the significance of threading template alignments and on the convergence parameters of the structure assembly simulations. The c-score ranges typically from −5 to 2, with higher scores indicating models with higher confidence. The obtained models were evaluated and visualized using the software PROCHECK [60], PyMol [61] MOLMOL [63] and Chimera [62]. Secondary structure estimation using the predicted models was performed using the software DSSP [64].
3.5. Molecular Dynamics
In order to garner insights into the stability of the modelled structure we run molecular dynamics simulation using Gromacs package (v.4.5.5) [42] with the Amber99 force field [65]. The ΔTM-FlmC protein model was centred in triclinic boxes allowing a 1 nm distance from each box edge and solvated by explicit solvent (TIP3P model), ending with 10493 water molecules. Counterions were randomly added to neutralize the system. First, the system was energy minimized using the steepest descent approach, followed by an equilibration phase where water molecules and protein heavy atoms were position restrained. The unrestrained systems were kept in an NPT ensemble, at constant temperature of 300 K by the velocity rescaling thermostat and at a pressure of 1 bar by the Berendsen thermostat. Electrostatic interactions were evaluated using the particle mesh Ewald method [66] and Lennard-Jones forces by a cut-off radius of 0.9 nm. Bond lengths involving hydrogen were restrained by the LINCS algorithm [67]. The time step was set to 2 fs and periodic boundary conditions were applied in all three dimensions. Protein structure simulation were run with no Mg (II) ion added.
Production run was carried for 5 ns of simulation time and the last frame of the trajectory has been used for the successive structural analyses.
Specifically, predictions of the binding pocket and docking studies were carried out on the final protein conformation of the MD simulation (rmsd = 0.4 nm on all atoms along the 5 ns simulation time).
3.6. ICP-MS
Mg(II) ion concentrations were measured via ICP-MS Agilent 7500ce (Agilent Technologies, Inc., USA), equipped with ORS tech interference/reaction cell to reduce polyatomic interferences. Instrument performances were checked using proper Tuning Solution (AGILENT® until the setting related to sensitivity and interference parameters were optimized. Interferences were tested through Interference Check Solutions (AGILENT®) in order to check the efficient functioning of ORS system.
Instrumental drift was monitored in continuum using Y-Tb internal standard with constant concentration. The analytical precision and accuracy for repeated quantifications of sample solution, international and internal standards (Agilent solutions EPA 200.8 Validated Standards, Agilent Technologies, Inc., USA), were better than 10%. Detected concentrations exceeded by at least one order of magnitude the limits of detection (LOD) and quantification (LOQ), according to Long and Winefordner [68]. Experiments were conducted in triplicate and for each sample different dilutions were analysed.
3.7. Lipid Extraction
The recombinant ΔTM-FlmC solution (20 mM sodium phosphate and 0.2 M NaCl at pH 6.8) was acidified with HCl in order to completely unfold the protein. The solution was then filtered using Amicon filters (Merck KGaA, Darmstadt, Germany) and the flow through was mixed with CH3Cl and CH3OH to obtain a final ratio of 2:2:1. The sample was incubated under stirring for 2 h. After centrifugation at 3750× g for 10 min, the lower organic fraction was recovered and dried. The resulting extract has been then analysed via TLC using an aluminium sheet Silica gel 60 F254 (Merck KGaA, Darmstadt, Germany) developed in a solution of CHCl3-CH3OH-H2O (13:7:2 ratio), sprayed with a solution of H2SO4-CH3COOH-H2O (1:20:4 ratio) and charred to visualize the organic compound.
3.8. UHPLC-HRMS Analyses
A Shimadzu NEXERA UHPLC system (Shimadzu, Japan) was used with a Luna® Omega C18 column (1.6 μm particle size, 50 × 2.1 mm i.d., Phenomenex, Torrance, CA, USA).
Separation was achieved with a linear gradient of water (A) and acetonitrile (B), both with 0.1% formic acid. Gradient conditions were as follows: 0–5 min, linear from 5% to 55% B; 5–10 min, linear from 55% to 75% B; 10–11 min, linear from 75% to 95% B; 11–13 min, isocratic 95% B. Then, the starting conditions were restored and the column was allowed to re-equilibrate for 1 min. The total run time was 14 min, with a flow rate of 0.5 mL min−1 and an injection volume of 2.0 μL.
MS analysis was performed using a hybrid QqTOF MS instrument, the AB SCIEX TripleTOF® 4600 (AB Sciex, Concord, ON, Canada), equipped with a DuoSprayTM ion source (consisting of both electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) probes), which was operated in positive and negative ionization modes. The instrument was controlled by Analyst® TF 1.7 software, while data processing was carried out using PeakView® software version 2.2.
4. Conclusions
The inspiring hypothesis of this study proposes the FlmA, FlmB and FlmC proteins from L. plantarum as belonging to the LytR-CpsA-Psr (LCP) protein family, a class of proteins widely distributed among Gram positive bacteria and involved in cell envelop biogenesis and homeostasis [37,69]. Members of the LCP protein family were initially indicated as transcriptional attenuators, due to the pleiotropic effects of null mutations in the corresponding genes but structural and biochemical analyses of LCP proteins from different Gram-positive species support the hypothesis of their role as transporters of anionic polymers, including wall teichoic acids, to the cell wall peptidoglycan [37,70].
The analysis of the Flm proteins indicated a high amino acid sequence identity with Cps2A and BrpA (Biofilm regulatory protein A) of Streptococcus mutans, both members of the LCP protein family [31,35,36]. In particular, in this study, among the three proteins we chose to characterize FlmC, due to the interesting compromised biofilm development and increased autolytic activity phenotypes, shown by the L. plantarum LM3-6 (ΔflmC) strain. Mutant strains carrying deletions in the other flm genes were only hampered in biofilm development, retaining a wild type autolytic activity [30]. These phenotypes were also found in S. mutants strains, carrying single null mutations in brpA or psr genes, while the deficiency in both genes resulted to be lethal in the conditions used for mutant selection [36].
Here we report the characterization of the FlmC protein that reveals the presence at the N-terminus of a hydrophobic fragment, likely a trans-membrane domain and at the C-terminus of a hydrophilic domain that we identify as belonging to the characterizing domain of the LytR-CpsA-Psr protein family. To validate our hypothesis, we have identified the portion of the L. plantarum FlmC protein to express, namely ΔTM-FlmC, for structural characterization. ΔTM-FlmC structural model has been then compared to the X-ray structure of the Cps2A protein obtained by Kaway and co-workers [37], evidencing how, apart from minor structural rearrangements, the two proteins appear highly similar. In particular, the two proteins share a hydrophobic cavity that in Cps2A has been proved to host a lipid molecule. Besides that, Cps2A, thanks to two crucial aspartate residues, binds octahedrally to a Mg(II) ion fundamental for its phosphatase activity [37]; the same residues are conserved in ΔTM-FlmC and show in the model a position compatible with the coordination of Mg(II). Accordingly, our experimental data have proved the co–purification with ΔTM-FlmC of a lipid molecule and the presence of a Mg(II) ion.
The cell wall is of great interest for its role in the communication with the environment, in the defence from a plethora of insults, in the localization of cell-surface expressed virulence factors and in biofilm development. Studies on Lactobacilli biofilms will help to shed light on their involvement in prevention of diseases related to disbiosis conditions, in particular in vaginal or intestinal environments [4,15,16,18]. Our results strongly suggest that the L. plantarum FlmC protein is a phosphotransferase, just as the well characterized Cps2A, supporting previous results demonstrating the role of this protein in biofilm development and autolysis activity.
Supplementary Materials
The following are available online, Figure S1: Binding site interactions analysys, Figure S2: UHPLC-HRMS analysis, Figure S3: ICP-MS analysis.
Author Contributions
G.D.A., A.P., R.M. and V.V. produced and purified samples and performed all the in-silico analysis. E.C. performed the ICP-MS experiments. S.P. (Severina Pacifico) and S.P. (Simona Piccolella) performed UHPLC-HRMS analyses. M.S., C.I., L.M. and G.M. designed experiments, conceived and supervised the study. G.D.A., L.M. and G.M. designed the study and wrote the manuscript.
Funding
This work was financially supported by the Italian MiUR (PRIN 20157WZM8A to G.M.)
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mihai, M.M.; Holban, A.M.; Giurcaneanu, C.; Popa, L.G.; Oanea, R.M.; Lazar, V.; Chifiriuc, M.C.; Popa, M.; Popa, M.I. Microbial biofilms: Impact on the pathogenesis of periodontitis, cystic fibrosis, chronic wounds and medical device-related infections. Curr. Top. Med. Chem. 2015, 15, 1552–1576. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Mangwani, N.; Kumari, S.; Das, S. Bacterial biofilms and quorum sensing: Fidelity in bioremediation technology. Biotechnol. Genet. Eng. Rev. 2016, 32, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Jagmann, N.; Philipp, B. Reprint of design of synthetic microbial communities for biotechnological production processes. J. Biotechnol. 2014, 192, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Nancharaiah, Y.V.; Venkata Mohan, S.; Lens, P.N. Metals removal and recovery in bioelectrochemical systems: A review. Bioresour. Technol. 2015, 195, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Lagrafeuille, R.; Souweine, B.; Forestier, C. Anti-biofilm activity as a health issue. Front. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Malgieri, G.; Avitabile, C.; Palmieri, M.; D’Andrea, L.D.; Isernia, C.; Romanelli, A.; Fattorusso, R. Structural basis of a temporin 1b analogue antimicrobial activity against gram negative bacteria determined by cd and nmr techniques in cellular environment. ACS Chem. Biol. 2015, 10, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Iacovino, R.; Caso, J.V.; Rapuano, F.; Russo, A.; Isidori, M.; Lavorgna, M.; Malgieri, G.; Isernia, C. Physicochemical characterization and cytotoxic activity evaluation of hydroxymethylferrocene: β-cyclodextrin inclusion complex. Molecules 2012, 17, 6056–6070. [Google Scholar] [CrossRef] [PubMed]
- Iacovino, R.; Rapuano, F.; Caso, J.V.; Russo, A.; Lavorgna, M.; Russo, C.; Isidori, M.; Russo, L.; Malgieri, G.; Isernia, C. β-cyclodextrin inclusion complex to improve physicochemical properties of pipemidic acid: Characterization and bioactivity evaluation. Int. J. Mol. Sci. 2013, 14, 13022–13041. [Google Scholar] [CrossRef] [PubMed]
- Avitabile, C.; Netti, F.; Orefice, G.; Palmieri, M.; Nocerino, N.; Malgieri, G.; D’Andrea, L.D.; Capparelli, R.; Fattorusso, R.; Romanelli, A. Design, structural and functional characterization of a temporin-1b analog active against gram-negative bacteria. Biochim. Biophys. Acta 2013, 1830, 3767–3775. [Google Scholar] [CrossRef] [PubMed]
- Bayramov, D.F.; Neff, J.A. Beyond conventional antibiotics—New directions for combination products to combat biofilm. Adv. Drug Deliv. Rev. 2017, 112, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Coleman, S.R.; Hancock, R.E. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr. Opin. Microbiol. 2016, 33, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Dillon, J.F. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 2007, 102, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Watters, C.; Fleming, D.; Bishop, D.; Rumbaugh, K.P. Host responses to biofilm. Prog. Mol. Biol Transl. Sci。 2016, 142, 193–239. [Google Scholar] [PubMed]
- Santos, C.M.; Pires, M.C.; Leão, T.L.; Hernández, Z.P.; Rodriguez, M.L.; Martins, A.K.; Miranda, L.S.; Martins, F.S.; Nicoli, J.R. Selection of Lactobacillus strains as potential probiotics for vaginitis treatment. Microbiology 2016, 162, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, E.; Adrain, V.; Diana, P. Microbial biofilm formation under the influence of various physical-chemical factors. Biotechnol. Biotechnol. Equip. 2010, 24, 1993–1996. [Google Scholar] [CrossRef]
- Leccese Terraf, M.C.; Juárez Tomás, M.S.; Rault, L.; Le Loir, Y.; Even, S.; Nader-Macías, M.E. Biofilms of vaginal Lactobacillus reuteri CRL 1324 and Lactobacillus rhamnosus CRL 1332: Kinetics of formation and matrix characterization. Arch. Microbiol. 2016, 198, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Aoudia, N.; Rieu, A.; Briandet, R.; Deschamps, J.; Chluba, J.; Jego, G.; Garrido, C.; Guzzo, J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016, 53, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Lönnermark, E.; Friman, V.; Lappas, G.; Sandberg, T.; Berggren, A.; Adlerberth, I. Intake of Lactobacillus plantarum reduces certain gastrointestinal symptoms during treatment with antibiotics. J. Clin. Gastroenterol. 2010, 44, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.C.; Caggianiello, G.; van Swam, I.I.; Taverne, N.; Meijerink, M.; Bron, P.A.; Spano, G.; Kleerebezem, M. Strain-specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host interactions. Appl. Environ. Microbiol. 2016, 82, 3959–3970. [Google Scholar] [CrossRef] [PubMed]
- Vicariotto, F.; Mogna, L.; Del Piano, M. Effectiveness of the two microorganisms Lactobacillus fermentum LF15 and Lactobacillus plantarum LP01, formulated in slow-release vaginal tablets, in women affected by bacterial vaginosis: A pilot study. J. Clin. Gastroenterol. 2014, 48, S106–S112. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; Wilson, G. Probiotics genomics. Microb. Biotechnol. 2010, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; van Hylckama Vlieg, J.E. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 2011. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Arena, M.P.; Fiocco, D.; Capozzi, V.; Drider, D.; Spano, G. Lactobacillus plantarum with broad antifungal activity: A promising approach to increase safety and shelf-life of cereal-based products. Int. J. Food Microbiol. 2017, 247, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, C.; Vastano, V.; Siciliano, R.A.; Candela, M.; Vici, M.; Muscariello, L.; Marasco, R.; Sacco, M. Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb. Cell Factor. 2009. [Google Scholar] [CrossRef] [PubMed]
- Vastano, V.; Capri, U.; Candela, M.; Siciliano, R.A.; Russo, L.; Renda, M.; Sacco, M. Identification of binding sites of Lactobacillus plantarum enolase involved in the interaction with human plasminogen. Microbiol. Res. 2013, 168, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Salzillo, M.; Vastano, V.; Capri, U.; Muscariello, L.; Sacco, M.; Marasco, R. Identification and characterization of enolase as a collagen-binding protein in Lactobacillus plantarum. J. Basic Microbiol. 2015, 55, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Vastano, V.; Pagano, A.; Fusco, A.; Merola, G.; Sacco, M.; Donnarumma, G. The Lactobacillus plantarum enoA1 enolase is involved in immunostimulation of caco-2 cells and in biofilm development. Adv. Exp. Med. Biol. 2016, 897, 33–44. [Google Scholar] [PubMed]
- Muscariello, L.; Marino, C.; Capri, U.; Vastano, V.; Marasco, R.; Sacco, M. CcpA and three newly identified proteins are involved in biofilm development in Lactobacillus plantarum. J. Basic Microbiol. 2013, 53, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Chatfield, C.H.; Koo, H.; Quivey, R.G. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 2005, 151, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Patras, K.A.; Derieux, J.; Al-Bassam, M.M.; Adiletta, N.; Vrbanac, A.; Lapek, J.D.; Zengler, K.; Gonzalez, D.J.; Nizet, V. Group b Streptococcus biofilm regulatory protein a contributes to bacterial physiology and innate immune resistance. J. Infect. Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.T.; Scott-Anne, K.; Liao, S.; De, A.; Luo, M.; Kovacs, C.; Narvaez, B.S.; Faustoferri, R.C.; Yu, Q.; Taylor, C.M.; et al. Deficiency of BrpA in Streptococcus mutans reduces virulence in rat caries model. Mol. Oral Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.T.; Baker, H.V.; Burne, R.A. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 2006, 188, 2983–2992. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, J.P.; Liao, S.; Yao, X.; Ahn, S.J.; Isoda, R.; Nguyen, A.H.; Brady, L.J.; Burne, R.A.; Abranches, J.; Wen, Z.T. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 2012, 78, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Bitoun, J.P.; Liao, S.; McKey, B.A.; Yao, X.; Fan, Y.; Abranches, J.; Beatty, W.L.; Wen, Z.T. Psr is involved in regulation of glucan production and double deficiency of BrpA and Psr is lethal in Streptococcus mutans. Microbiology 2013, 159, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Marles-Wright, J.; Cleverley, R.M.; Emmins, R.; Ishikawa, S.; Kuwano, M.; Heinz, N.; Bui, N.K.; Hoyland, C.N.; Ogasawara, N.; et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011, 30, 4931–4941. [Google Scholar] [CrossRef] [PubMed]
- Gräslund, S.; Nordlund, P.; Weigelt, J.; Hallberg, B.M.; Bray, J.; Gileadi, O.; Knapp, S.; Oppermann, U.; Arrowsmith, C.; Hui, R.; et al. Protein production and purification. Nat. Methods 2008, 5, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. Aggrescan: A server for the prediction and evaluation of “Hot spots” of aggregation in polypeptides. BMC Bioinform. 2007. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-tasser: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. Gromacs 4: Algorithms for highly efficient, load-balanced and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Kellis, J.T.; Nyberg, K.; Fersht, A.R. Energetics of complementary side-chain packing in a protein hydrophobic core. Biochemistry 1989, 28, 4914–4922. [Google Scholar] [CrossRef] [PubMed]
- Shortle, D.; Stites, W.E.; Meeker, A.K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry 1990, 29, 8033–8041. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Stigter, D. Modeling protein stability as heteropolymer collapse. Adv. Protein. Chem. 1995, 46, 59–104. [Google Scholar] [PubMed]
- Baglivo, I.; Palmieri, M.; Rivellino, A.; Netti, F.; Russo, L.; Esposito, S.; Iacovino, R.; Farina, B.; Isernia, C.; Fattorusso, R.; et al. Molecular strategies to replace the structural metal site in the prokaryotic zinc finger domain. Biochim. Biophys. Acta 2014, 1844, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Malgieri, G.; Eliezer, D. Structural effects of parkinson’s disease linked DJ-1 mutations. Protein Sci. 2008, 17, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Hendlich, M.; Rippmann, F.; Barnickel, G. Ligsite: Automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph. Model. 1997, 15, 359–363. [Google Scholar] [CrossRef]
- Wass, M.N.; Kelley, L.A.; Sternberg, M.J. 3dligandsite: Predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010, 38, W469–W473. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. Swissdock, a protein-small molecule docking web service based on eadock dss. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Park, H.; Heo, L.; Seok, C. Galaxyweb server for protein structure prediction and refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef] [PubMed]
- De Paola, I.; Pirone, L.; Palmieri, M.; Balasco, N.; Esposito, L.; Russo, L.; Mazzà, D.; Di Marcotullio, L.; Di Gaetano, S.; Malgieri, G.; et al. Cullin3-btb interface: A novel target for stapled peptides. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Spaziano, G.; Sorrentino, R.; Matteis, M.; Malgieri, G.; Sgambato, M.; Russo, T.P.; Terlizzi, M.; Roviezzo, F.; Rossi, F.; Pinto, A.; et al. Nociceptin reduces the inflammatory immune microenvironment in a conventional murine model of airway hyperresponsiveness. Clin. Exp. Allergy 2017, 47, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; Malgieri, G.; Russo, L.; Baglivo, I.; Esposito, S.; Netti, F.; Del Gatto, A.; de Paola, I.; Zaccaro, L.; Pedone, P.V.; et al. Structural Zn(ii) implies a switch from fully cooperative to partly downhill folding in highly homologous proteins. J. Am. Chem. Soc. 2013, 135, 5220–5228. [Google Scholar] [CrossRef] [PubMed]
- D′Abrosca, G.; Russo, L.; Palmieri, M.; Baglivo, I.; Netti, F.; de Paola, I.; Zaccaro, L.; Farina, B.; Iacovino, R.; Pedone, P.V.; et al. The (unusual) aspartic acid in the metal coordination sphere of the prokaryotic zinc finger domain. J. Inorg. Biochem. 2016, 161, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.; Lloyd, G.; Joe, M.; Lowary, T.L.; Reynolds, E.; Walters-Morgan, H.; Bhatt, A.; Lovering, A.; Besra, G.S.; Alderwick, L.J. Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. Am. Soc. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Paladino, A.; Zangi, R. Ribose 2′-hydroxyl groups stabilize RNA hairpin structures containing GCUAA pentaloop. J. Chem. Theory Comput. 2013, 9, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad Sci. USA 2015, 112, 3095–3103. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. Aqua and procheck-nmr: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool, 2nd ed.; CCP4 Newsletter on Protein Crystallography: San Carlos, CA, USA, 2002. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wüthrich, K. Molmol: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Joosten, R.P.; te Beek, T.A.; Krieger, E.; Hekkelman, M.L.; Hooft, R.W.; Schneider, R.; Sander, C.; Vriend, G. A series of pdb related databases for everyday needs. Nucleic. Acids Res. 2011, 39, D411–D419. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- York, D.M.; Wlodawer, A.; Pedersen, L.G.; Darden, T.A. Atomic-level accuracy in simulations of large protein crystals. Proc. Natl. Acad. Sci. USA 1994, 91, 8715–8718. [Google Scholar] [CrossRef] [PubMed]
- Hess, B. P-lincs: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Long, G.L.; Winefordner, J.D. Limit of detection a closer look at the iupac definition. Anal. Chem. 1983, 55, 712A–724A. [Google Scholar]
- Hübscher, J.; Lüthy, L.; Berger-Bächi, B.; Stutzmann Meier, P. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genet. 2008. [Google Scholar] [CrossRef] [PubMed]
- Siegel, S.D.; Liu, J.; Ton-That, H. Biogenesis of the gram-positive bacterial cell envelope. Curr. Opin. Microbiol. 2016, 34, 31–37. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).