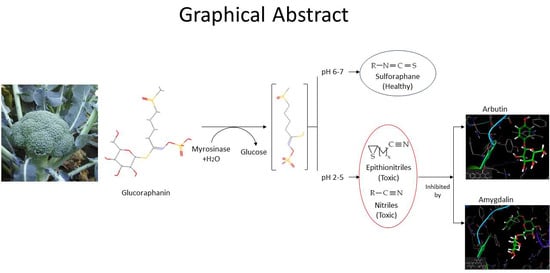
Molecular Docking of Potential Inhibitors of Broccoli Myrosinase
Abstract
:1. Introduction
2. Results
2.1. Effect of pH on Myrosinase Activity
2.2. Molecular Docking of Broccoli Myrosinase with Substrates and Potential Inhibitors
2.3. Molecular Interactions
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Myrosinase Activity
4.3. Effect of pH on Myrosinase Activity
4.4. Molecular Docking
4.4.1. Preparation of Broccoli Myrosinase Structure
4.4.2. Ligand Preparation
4.4.3. Receptor Grid Generation
4.4.4. Glide Standard Precision (SP) Ligand Docking
4.4.5. Residues that Stabilize Myrosinase-Ligand Complexes
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Martinez-Ballesta, M.D.; Moreno, D.A.; Carvajal, M. The Physiological Importance of Glucosinolates on Plant Response to Abiotic Stress in Brassica. Int. J. Mol. Sci. 2013, 14, 11607–11625. [Google Scholar] [CrossRef] [PubMed]
- Latte, K.P.; Appel, K.E.; Lampen, A. Health benefits and possible risks of broccoli—An overview. Food Chem. Toxicol. 2011, 49, 3287–3309. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, K.L. Chapter 40-Glucosinolates. In Nutraceuticals, 1st ed.; Gupta, R.C., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 551–554. [Google Scholar] [CrossRef]
- Bohinc, T.; Goreta Ban, S.; Ban, D.; Trdan, S. Glucosinolates in plant protection strategies: A review. Arch. Biol. Sci. 2012, 64, 821–828. [Google Scholar] [CrossRef] [Green Version]
- Holst, B.; Fenwick, G.R. Glucosinolates. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 2922–2930. ISBN 9780122270550. [Google Scholar] [CrossRef]
- Wittstock, U.; Meier, K.; Dörr, F.; Ravindran, B.M. NSP-Dependent Simple Nitrile Formation Dominates upon Breakdown of Major Aliphatic Glucosinolates in Roots, Seeds, and Seedlings of Arabidopsis thaliana Columbia. Front. Plant. Sci. 2016, 7, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Aires, A.; Mota, V.R.; Saavedra, M.J.; Rosa, E.A.; Bennett, R.N. The antimicrobial effects of glucosinolates and their respective enzymatic hydrolysis products on bacteria isolated from the human intestinal tract. J. Appl. Microbiol. 2009, 106, 2086–2095. [Google Scholar] [CrossRef] [PubMed]
- Vig, A.P.; Rampal, G.; Thind, T.S.; Arora, S. Bio-protective effects of glucosinolates—A review. LWT-Food Sci. Technol. 2009, 42, 1561–1572. [Google Scholar] [CrossRef]
- Holst, B.; Williamson, G. A critical review of the bioavailability of glucosinolates and related compounds. Nat. Prod. Rep. 2004, 21, 425–447. [Google Scholar] [CrossRef] [PubMed]
- Bartnik, M.; Facey, P.C. Chapter 8—Glycosides A2. In Pharmacognosy; Simone, B., Delgoda, R., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 101–161. [Google Scholar] [CrossRef]
- Hill, G.D. Plant Antinutritional Factors. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 4578–4587. ISBN 9780080917917. [Google Scholar]
- Jeffery, E.H.; Keck, A.S. Translating knowledge generated by epidemiological and in vitro studies into dietary cancer prevention. Mol. Nutr. Food Res. 2008, 52, S7–S17. [Google Scholar] [PubMed]
- Mays, J.R.; Weller Roska, R.L.; Sarfaraz, S.; Mukhtar, H.; Rajski, S.R. Identification, synthesis, and enzymology of non-natural glucosinolate chemopreventive candidates. ChemBioChem 2008, 9, 729–747. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.; Pun, S.; Chaliha, M.; Scheelings, P.; O’Hare, T. An unusual combination in papaya (Carica papaya): The good (glucosinolates) and the bad (cyanogenic glycosides). J. Food Compos. Anal. 2013, 29, 82–86. [Google Scholar] [CrossRef]
- Mahn, A.V.; Román, J.I.; Reyes, A.E. Effect of freeze-drying of pre-processed broccoli on drying kinetics and sulforaphane content. Inf. Technol. 2016, 27, 95–106. [Google Scholar] [CrossRef]
- Mahn, A.; Pérez, C. Optimization of an incubation step to maximize sulforaphane content in pre-processed broccoli. J. Food Sci. Technol. 2016, 53, 4110–4115. [Google Scholar] [CrossRef] [PubMed]
- Houghton, C.A.; Fassett, R.G.; Coombes, J.S. Sulforaphane: Translational research from laboratory bench to clinic. Nutr. Rev. 2013, 71, 709–726. [Google Scholar] [CrossRef] [PubMed]
- Mahn, A.; Angulo, A.; Cabañas, F. Purification and characterization of broccoli (Brassica oleracea var. italica) myrosinase (beta-thioglucosidase glucohydrolase). J. Agric. Food Chem. 2014, 62, 11666–11671. [Google Scholar] [CrossRef] [PubMed]
- Roman, J.; Castillo, A.; Cottet, L.; Mahn, A. Kinetic and structural study of broccoli myrosinase and its interaction with different glucosinolates. Food Chem. 2018, 254, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Stoin, D.; Pirsan, P.; Radu, F.; Poian, M.A.; Alexa, E.; Dogaru, D. Studies regarding the myrosinase enzymatic activity from black mustard (Brassica nigra) seeds. J. Food Agric. Environ. 2009, 7, 44–47. [Google Scholar]
- Martinez-Ballesta, M.D.; Carvajal, M. Myrosinase in Brassicaceae: The most important issue for glucosinolate turnover and food quality. Phytochem. Rev. 2015, 14, 1045–1051. [Google Scholar] [CrossRef]
- Cartea, M.E.; Velasco, P. Glucosinolates in Brassica foods: Bioavailability in food and significance for human health. Phytochem. Rev. 2008, 7, 213–229. [Google Scholar] [CrossRef]
- Van Eylen, D.; Oey, I.; Hendrickx, M.; Van Loey, A. Behavior of mustard seed (Sinapis alba L.) myrosinase during temperature/pressure treatments: A case study on enzymeactivity and stability. Eur. Food Res. Technol. 2008, 226, 545–553. [Google Scholar] [CrossRef]
- Ohtsuru, M.; Kawatani, H. Studies of the myrosinase from Wasabi japonica: Purification and some properties of wasabi myrosinase. Agric. Biol. Chem. 1979, 43, 2249–2255. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, S.; Sangwanc, S.; Yadava, I.S.; Yadav, R. Protein modeling and active site binding mode interactions of myrosinase–sinigrin in Brassica juncea—An in silico approach. J. Mol. Graph. Model. 2011, 29, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Cardoso, C.; Genta, F.; Terra, R.; Ferreira, C. Active site characterization and molecular cloning of Tenebrio molitor midgut trehalase and comments on their insect homologs. Insect Biochem. Mol. Biol. 2013, 43, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Blaheta, R.A.; Nelson, K.; Haferkamp, A.; Juengel, E. Amygdalin, quackery or cure? Phytomedicine 2016, 23, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Migas, P.; Krauze-Baranowska, M. The significance of arbutin and its derivatives in therapy and cosmetics. Phytochem. Lett. 2015, 13, 35–40. [Google Scholar] [CrossRef]
- Schrödinger Release 2018-1: Maestro; Schrödinger, LLC: New York, NY, USA, 2018.
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System; Version 2.0; Schrödinger, LLC: New York, NY, USA, 2018.
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-1: Schrödinger Suite 2018-1 Protein Preparation Wizard; Epik, Schrödinger, LLC: New York, NY, USA, 2016; Impact, Schrödinger, LLC: New York, NY, USA, 2016; Prime, Schrödinger, LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: LigPrep; Schrödinger, LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: Schrödinger Suite 2018-1 Induced Fit Docking protocol; Glide, Schrödinger, LLC: New York, NY, USA, 2016; Prime, Schrödinger, LLC: New York, NY, USA, 2018.
Sample Availability: Samples of the compounds 1–40 are not available from the authors. |








| N° | Inhibitor | PubChem CID * | Glide Score (kcal/mol) | Docking Score |
|---|---|---|---|---|
| 1 | (2-sulfate) ethyl 1-thio-beta-d-glucopyranoside | 23666133 | −6.152 | −6.152 |
| 2 | (2R,5R)-dihydroxymethyl-(3R,4R)-dihydroxypyrrolidine | 124702 | −5.167 | −5.167 |
| 3 | (3-sulfonate) propyl 1-thio-beta-d-glucopyranoside | 23674091 | −6.392 | −6.392 |
| 4 | 1,10-phenanthroline | 1318 | −5.788 | −5.782 |
| 5 | 1,4-dideoxy-1,4-imino-d-arabinitol | 451991 | −5.508 | −5.508 |
| 6 | 1-deoxynojirimycin | 29435 | −4.671 | −4.671 |
| 7 | 1-O-methyl-alpha-d-glucopyranose | 64947 | −5.372 | −5.372 |
| 8 | 2-methoxy-5-nitrotropone | 84563 | −5.774 | −5.148 |
| 9 | 5,5′-dithiobis (2-nitrobenzoic acid) | 6254 | −6.076 | −5.984 |
| 10 | Alexine | 189,377 | −4.778 | −4.778 |
| 11 | Amygdalin | 656,516 | −6.964 (−6.900) −6.918 (−6.600) | −6.918 |
| 12 | Arbutin | 440,936 | −7.842 (−6.900) −7.474 (−6.500) | −7.474 |
| 13 | l-ascorbic acid | 54,670,067 | −5.626 | −4.682 |
| 14 | Castanospermine | 54,445 | −6.426 | −6.426 |
| 15 | Cysteamine | 6058 | −2.981 | −2.981 |
| 16 | Delta-gluconolactone | 7027 | −5.566 | −5.566 |
| 17 | Diisopropyl fluorophosphate | 5936 | −4.601 | −4.601 |
| 18 | Dithiothreitol | 446,094 | −3.613 | −3.613 |
| 19 | Ethylenediaminetetraacetic acid | 6049 | −2.266 | −2.170 |
| 20 | Fluorodinitrobenzene | 6264 | −5.331 | −5.331 |
| 21 | Fructose | 5984 | −4.356 | −4.356 |
| 22 | Galactose | 6036 | −5.671 | −5.671 |
| 23 | Glucose | 64,689 | −6.156 | −6.156 |
| 24 | l-Cysteine | 5862 | −4.176 | −4.127 |
| 25 | Maltose | 6255 | −5.396 | −5.396 |
| 26 | Mannose | 18,950 | −6.239 | −6.239 |
| 27 | Methyl jasmonate | 5,281,929 | −4.966 | −4.966 |
| 28 | Methyl-beta-d-glucopyranoside | 445,238 | −5.787 | −5.787 |
| 29 | Monochlorotrifluoro-p-benzoquinone | 53,662,935 | −5.092 | −5.092 |
| 30 | p-diazabenzenesulfonic acid | 67,540 | −4.707 | −4.594 |
| 31 | p-nitrophenyl-beta-d-glucopyranoside | 92,930 | −5.842 | −5.842 |
| 32 | Phenyl-beta-d-glucopyranoside | 65,080 | −6.524 | −6.524 |
| 33 | Salicin | 439,503 | −5.950 | −5.950 |
| 34 | Sorbitol | 5780 | −4.013 | −4.013 |
| 35 | Sucrose | 5988 | −5.305 | −5.305 |
| 36 | Thiobenzoate | 80,024 | −5.088 | −5.088 |
| 37 | Thiomalate | 5,352,130 | −5.475 | −4.782 |
| 38 | Thiophenol | 7969 | −4.786 | −4.786 |
| 39 | Trinitrobenzenesulfonic acid | 11,045 | −5.470 | −5.470 |
| 40 | Xylose | 135,191 | −6.164 | −6.164 |
| Mean glide and docking score | --- | −5.341 | −5.276 | |
| Sinigrin | 23,682,211 | −5.508 | −5.508 | |
| Glucoraphanin | 9,548,634 | −6.649 | −6.649 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Román, J.; Castillo, A.; Mahn, A. Molecular Docking of Potential Inhibitors of Broccoli Myrosinase. Molecules 2018, 23, 1313. https://doi.org/10.3390/molecules23061313
Román J, Castillo A, Mahn A. Molecular Docking of Potential Inhibitors of Broccoli Myrosinase. Molecules. 2018; 23(6):1313. https://doi.org/10.3390/molecules23061313
Chicago/Turabian StyleRomán, J., A. Castillo, and A. Mahn. 2018. "Molecular Docking of Potential Inhibitors of Broccoli Myrosinase" Molecules 23, no. 6: 1313. https://doi.org/10.3390/molecules23061313