Comparative in Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications
Abstract
:1. Introduction
2. Results
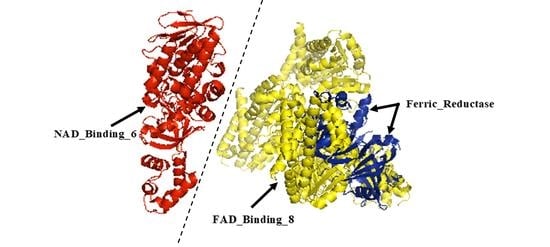
2.1. Identification, Classification, and Annotation of FRO Family Members
2.2. Systematic Evolutionary Relationship, Gene Structural Diversity, and Motif Analysis
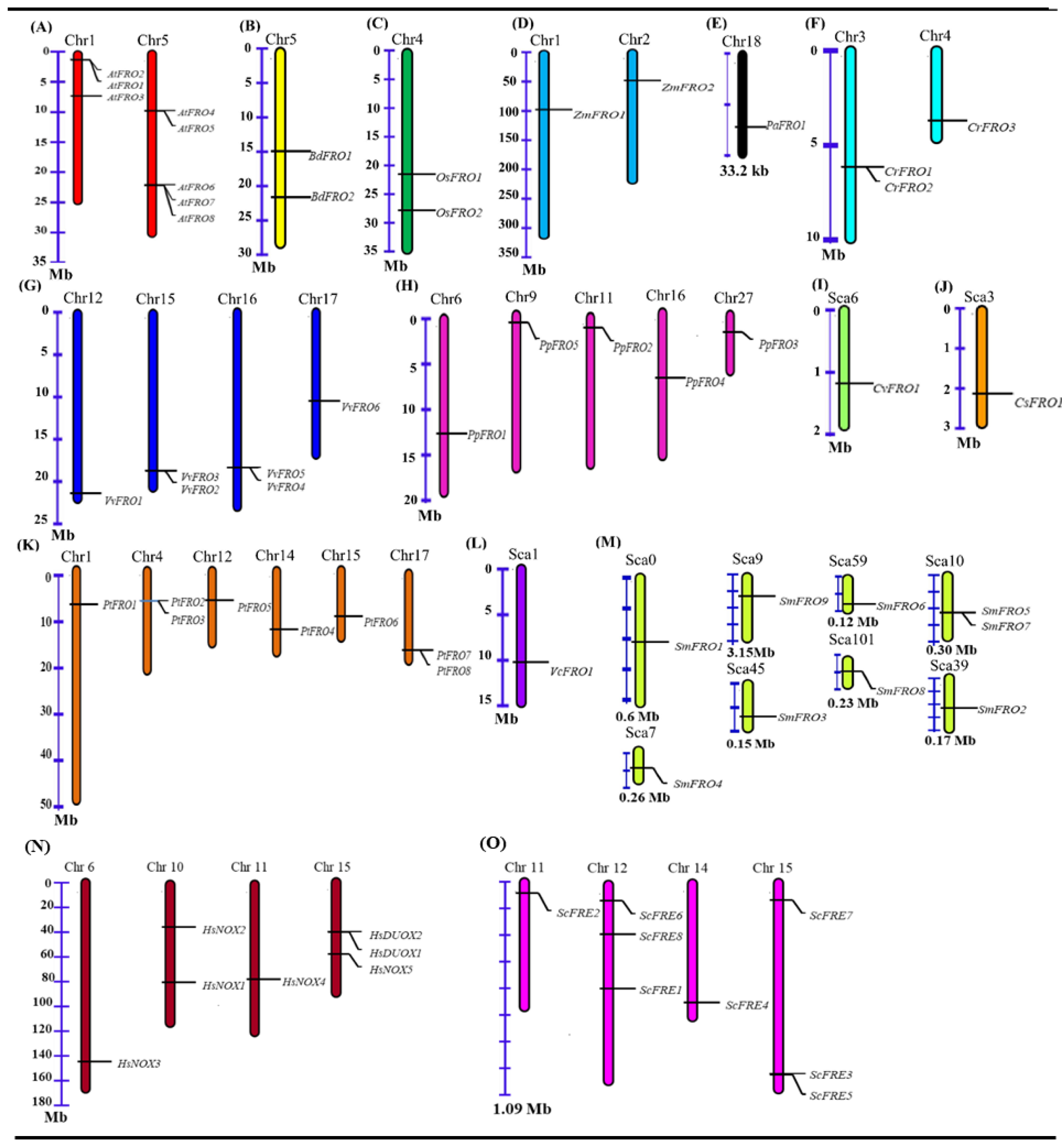
2.3. Chromosomal Location and Gene Duplication of FRO Genes
2.4. Developmental and Tissue-Specific Expression Profile of Rice OsFRO1 and OsFRO7 Genes
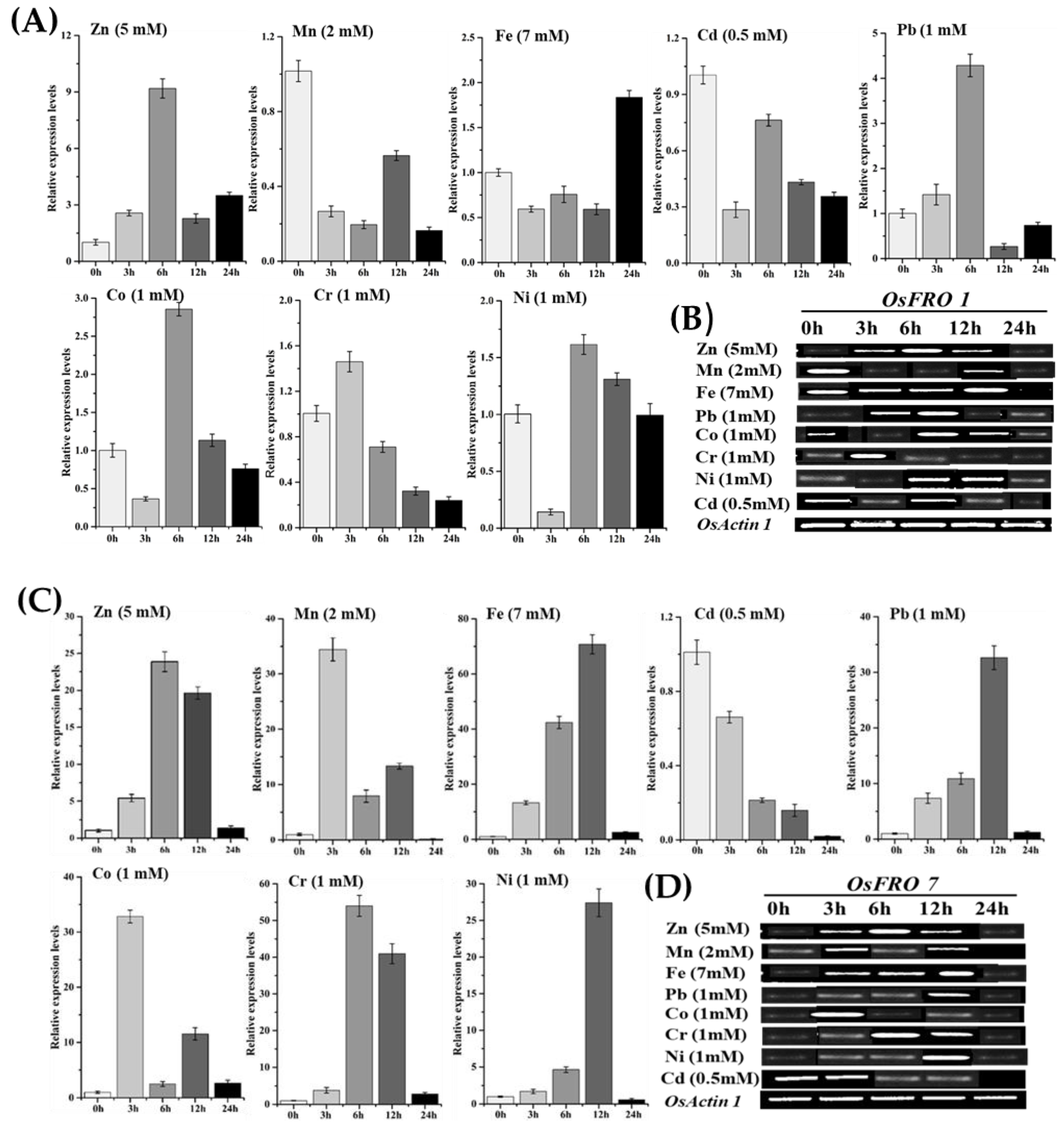
2.5. Inducible Expression Analysis of Rice OsFRO1 and OsFRO7 Gene under Abiotic and Hormones Applications
2.6. Inducible Expression Analysis of Rice OsFRO1 and OsFRO7 Genes against Different Heavy Metals Stresses
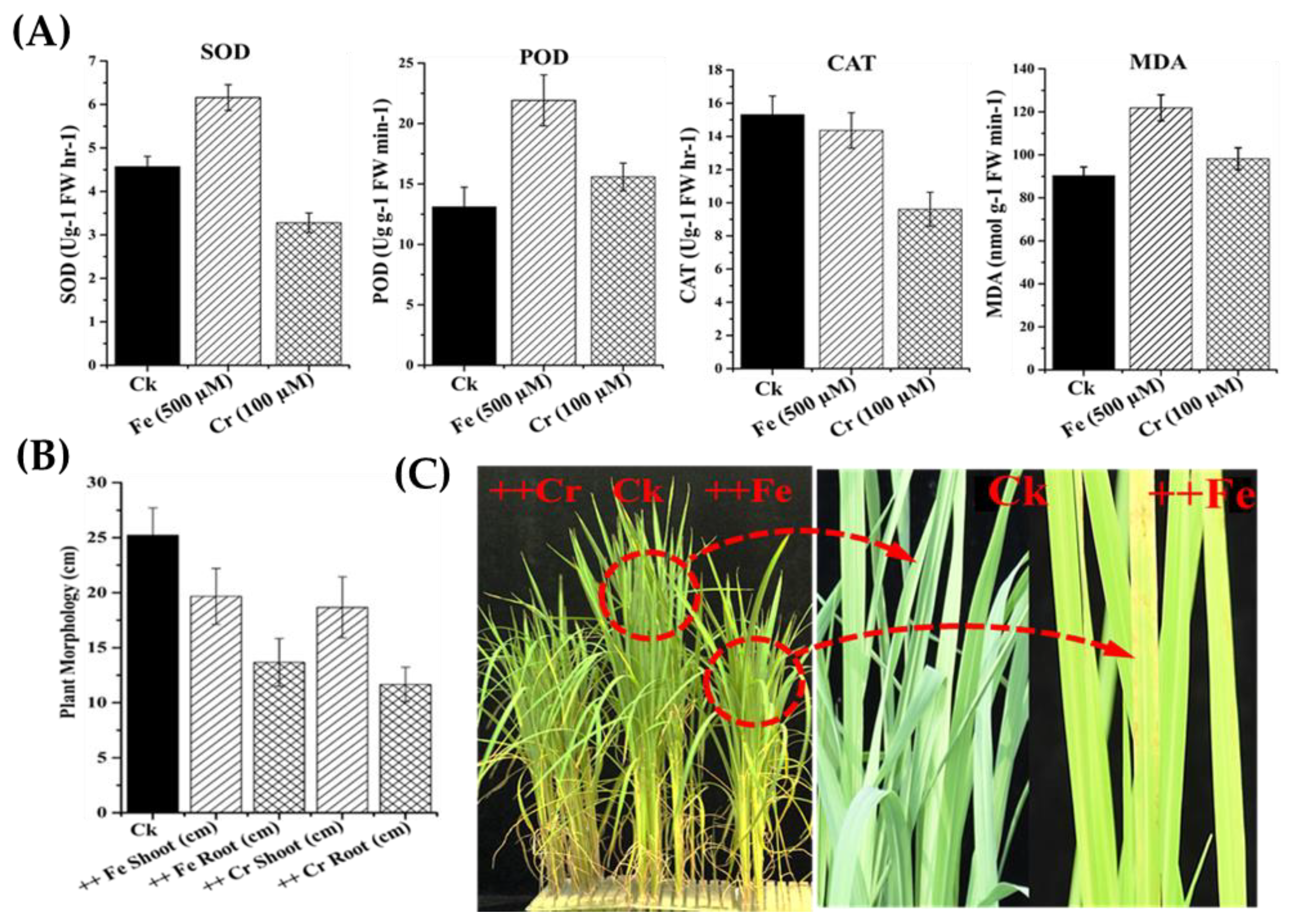
2.7. Determination of Antioxidant Enzyme Activities
3. Discussion
3.1. FRO Genes Expansion, Duplication, and Structural Diversity
3.2. Evolution and Functional Diversity of Conserved Domains in Plant FRO Genes
3.3. Tissue-Specific Gene Expression of Rice OsFRO1 and OsFRO7 and Specific Role in Response to Metals Stress
3.4. Inducible Expression Pattern of OsFRO1 and OsFRO7 Gene against Environmental Stresses and Hormones Treatments
3.5. Changes in Antioxidant Activity under Iron (Fe) and Chromium (Cr) Stress
4. Materials and Methods
4.1. Sequences Assembly and Identification of FRO Genes Family Members
4.2. Phylogenetic Relationships, Exon and Intron Distribution and Conserved Motif Analysis
4.3. Determining Chromosomal Location and Gene Duplication
4.4. Cis-Regulatory Elements in the Promoter Region of FRO Genes
4.5. Plant Material and Growth Conditions
4.6. Stress Treatments and Sample Collection
4.7. Antioxidant Enzyme Activities under Metal Stresses
4.8. qRT-PCR Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Victoria, F.D.C.; Bervald, C.M.P.; da Maia, L.C.; de Sousa, R.O.; Panaud, O.; de Oliveira, A.C. Phylogenetic relationships and selective pressure on gene families related to iron homeostasis in land plants. Genome 2012, 55, 883–900. [Google Scholar] [CrossRef] [PubMed]
- Takagi, S.I.; Nomoto, K.; Takemoto, T. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J. Plant Nutr. 1984, 7, 469–477. [Google Scholar] [CrossRef]
- Grotz, N.; Guerinot, M.L. Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2006, 1763, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Bashir, K.; Inoue, H.; Takahashi, M.; Nakanishi, H.; Nishizawa, N.K. Accumulation of starch in Zn-deficient rice. Rice 2012, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Vigani, G.; Morandini, P.; Murgia, I. Searching iron sensors in plants by exploring the link among 2′-OG-dependent dioxygenases, the iron deficiency response and metabolic adjustments occurring under iron deficiency. Front. Plant Sci. 2013, 4, 169. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Rasheed, S.; Kobayashi, T.; Seki, M.; Nishizawa, N.K. Regulating subcellular metal homeostasis: The key to crop improvement. Front. Plant Sci. 2016, 7, 1192. [Google Scholar] [CrossRef] [PubMed]
- Quinet, M.; Vromman, D.; Clippe, A.; Bertin, P.; Lequeux, H.; Dufey, I.; Lutts, S.; Lefevre, I. Combined transcriptomic and physiological approaches reveal strong differences between short-and long-term response of rice (Oryza sativa) to iron toxicity. Plant Cell Environ. 2012, 35, 1837–1859. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Ishimaru, Y.; Nishizawa, N.K. Molecular mechanisms of zinc uptake and translocation in rice. Plant Soil 2012, 361, 189–201. [Google Scholar] [CrossRef]
- Bashir, K.; Nishizawa, N.K. Iron proteins, plant iron transporters. In Encyclopedia of Metalloproteins; Springer: New York, NY, USA, 2013; pp. 1015–1023. [Google Scholar]
- Finazzi, G.; Petroutsos, D.; Tomizioli, M.; Flori, S.; Sautron, E.; Villanova, V.; Rolland, N.; Seigneurin-Berny, D. Ions channels/transporters and chloroplast regulation. Cell Calcium 2015, 58, 86–97. [Google Scholar] [CrossRef] [PubMed]
- López-Millán, A.F.; Duy, D.; Philippar, K. Chloroplast iron transport proteins–function and impact on plant physiology. Front. Plant Sci. 2016, 7, 178. [Google Scholar] [CrossRef] [PubMed]
- Finatto, T.; de Oliveira, A.C.; Chaparro, C.; Da Maia, L.C.; Farias, D.R.; Woyann, L.G.; Mistura, C.C.; Soares-Bresolin, A.P.; Llauro, C.; Panaud, O. Abiotic stress and genome dynamics: Specific genes and transposable elements response to iron excess in rice. Rice 2015, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Connolly, E.L. Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Sci. 2009, 176, 709–714. [Google Scholar] [CrossRef]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Schagerlöf, U.; Wilson, G.; Hebert, H.; Al-Karadaghi, S.; Hägerhäll, C. Transmembrane topology of FRO2, a ferric chelate reductase from Arabidopsis thaliana. Plant Mol. Biol. 2006, 62, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-F.; Li, W.-Q.; Li, W.-Y.; Wu, G.-L.; Zhou, C.-Y.; Chen, K.-M. Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 2013, 14, 9440–9458. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Bedard, K.; Lardy, B.; Krause, K.-H. NOX family NADPH oxidases: Not just in mammals. Biochimie 2007, 89, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, J.; Ríos-Momberg, M.; Hewitt, D.; Hansberg, W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005, 13, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.; Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 2007, 19, 4022–4034. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, L.; Du, J.; Yuan, Y.; Cheng, X.; Ling, H.-Q. Molecular and biochemical characterization of the Fe (III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Blevins, D.G.; Eide, D.J. Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 2002, 129, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cheng, X.; Ling, H.-Q. Isolation and characterization of Fe (III)-chelate reductase gene LeFRO1 in tomato. Plant Mol. Biol. 2004, 54, 125–136. [Google Scholar] [PubMed]
- Chang, Y.-L.; Li, W.-Y.; Miao, H.; Yang, S.-Q.; Li, R.; Wang, X.; Li, W.-Q.; Chen, K.-M. Comprehensive genomic analysis and expression profiling of the NOX gene families under abiotic stresses and hormones in plants. Genome Biol. Evol. 2016, 8, 791–810. [Google Scholar] [CrossRef] [PubMed]
- Gama, F.; Saavedra, T.; Dandlen, S.; de Varennes, A.; Correia, P.J.; Pestana, M.; Nolasco, G. Silencing of the FRO1 gene and its effects on iron partition in Nicotiana benthamiana. Plant Physiol. Biochem. 2017, 114, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.L.; Campbell, N.H.; Grotz, N.; Prichard, C.L.; Guerinot, M.L. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003, 133, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, I.; Campbell, N.H.; Ash, J.S.; Connolly, E.L. Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 2006, 223, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Guerinot, M.L. Homing in on iron homeostasis in plants. Trends Plant Sci. 2009, 14, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Hindt, M.N.; Guerinot, M.L. Getting a sense for signals: Regulation of the plant iron deficiency response. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2012, 1823, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Wilson, G.T.; Connolly, E.L. The diverse roles of FRO family metalloreductases in iron and copper homeostasis. Front. Plant Sci. 2014, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Ruengphayak, S.; Ruanjaichon, V.; Saensuk, C.; Phromphan, S.; Tragoonrung, S.; Kongkachuichai, R.; Vanavichit, A. Forward screening for seedling tolerance to Fe toxicity reveals a polymorphic mutation in ferric chelate reductase in rice. Rice 2015, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Lucena, C.; Romera, F.J.; Alcántara, E.; Pérez-Vicente, R. Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 2010, 61, 3885–3899. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, G.; Perrone, S.; Tataranno, M.L. Oxygen Toxicity: Chemistry and Biology of Reactive Oxygen Species. Semin. Fetal Neonatal Med. 2010, 15, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Kehrer, J.P. The Haber–Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2012, 41, D1152–D1158. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Dat, J.; Vandenabeele, S.; Vranová, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. CMLS 2000, 57, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.J.; Zhang, B.; Shi, W.W.; Li, H.Y. Hydrogen peroxide in plants: A versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 2008, 50, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, A.; Karen, S.; Jos, R.; Kelly, O.; Els, K.; Tony, R.; Nele, H.; Nathalie, V.; Yves, G.; Jan, C. The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 2011, 168, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Morgante, M.; Brunner, S.; Pea, G.; Fengler, K.; Zuccolo, A.; Rafalski, A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005, 37, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010, 20, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Sperotto, R.A.; Boff, T.; Duarte, G.L.; Santos, L.S.; Grusak, M.A.; Fett, J.P. Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J. Plant Physiol. 2010, 167, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.; Stein, R.J.; Fett-Neto, A.G.; Fett, J.P. Iron homeostasis related genes in rice. Genet. Mol. Biol. 2003, 26, 477–497. [Google Scholar] [CrossRef]
- Zhang, Q.; Jia, M.; Xing, Y.; Qin, L.; Li, B.; Jia, W. Genome-wide identification and expression analysis of MRLK family genes associated with strawberry (Fragaria vesca) fruit ripening and abiotic stress responses. PLoS ONE 2016, 11, e0163647. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kishino, H. Genomic background predicts the fate of duplicated genes: Evidence from the yeast genome. Genetics 2004, 166, 1995–1999. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, B.; Yu, L.; Feng, D.; Wang, H.; Wang, J. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol. Biol. 2009, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Krause, K.-H.; Xenarios, I.; Soldati, T.; Boeckmann, B. Evolution of the ferric reductase domain (FRD) superfamily: Modularity, functional diversification, and signature motifs. PLoS ONE 2013, 8, e58126. [Google Scholar] [CrossRef] [PubMed]
- Finegold, A.A.; Shatwell, K.P.; Segal, A.W.; Klausner, R.D.; Dancis, A. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 1996, 271, 31021–31024. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, M.; Eckert, H.; Arahana, V.; Graef, G.; Grusak, M.A.; Clemente, T. Molecular and phenotypic characterization of transgenic soybean expressing the Arabidopsis ferric chelate reductase gene, FRO2. Planta 2006, 224, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008, 275, 3249–3277. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Rivera, M.C.; Lake, J.A. Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. USA 1999, 96, 3801–3806. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Mekhedov, S.L.; Mirkin, B.G.; Koonin, E.V. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005, 33, 4626–4638. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Makarova, K.S.; Mekhedov, S.L.; Wolf, Y.I.; Koonin, E.V. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 2008, 25, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Kakei, Y.; Shimo, H.; Bashir, K.; Sato, Y.; Sato, Y.; Uozumi, N.; Nakanishi, H.; Nishizawa, N.K. A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J. Biol. Chem. 2011, 286, 24649–24655. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Senoura, T.; Sugimoto, K.; Ono, K.; Yano, M.; Ishikawa, S.; Arao, T. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef] [PubMed]
- Kakei, Y.; Ishimaru, Y.; Kobayashi, T.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. OsYSL16 plays a role in the allocation of iron. Plant Mol. Biol. 2012, 79, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Hervé, C.; Tonon, T.; Collén, J.; Corre, E.; Boyen, C. NADPH oxidases in Eukaryotes: Red algae provide new hints! Curr. Genet. 2006, 49, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.; Bothwell, J.H.; Laohavisit, A.; Smith, A.G.; Davies, J.M. NOX or not? Evidence for algal NADPH oxidases. Trends Plant Sci. 2011, 16, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Ricachenevsky, F.K.; Sperotto, R.A. There and back again, or always there? The evolution of rice combined strategy for Fe uptake. Front. Plant Sci. 2014, 5, 189. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Hanada, K.; Shimizu, M.; Seki, M.; Nakanishi, H.; Nishizawa, N.K. Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 2014, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, N.; Beyene, G.; Chauhan, R.D.; Gaitán-Solis, E.; Grusak, M.A.; Taylor, N.; Anderson, P. Overexpression of Arabidopsis VIT1 increases accumulation of iron in cassava roots and stems. Plant Sci. 2015, 240, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Vigani, G. Discovering the role of mitochondria in the iron deficiency-induced metabolic responses of plants. J. Plant Physiol. 2012, 169, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kabir, A. Biochemical and molecular changes in rice seedlings (Oryza sativa L.) to cope with chromium stress. Plant Biol. 2016, 18, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Hussain, K.; Nisar, M.F.; Majeed, A.; Nawaz, K.; Bhatti, K.H.; Afghan, S.; Shahazad, A.; Zia-ul-Hussnian, S. What molecular mechanism is adapted by plants during salt stress tolerance? Afr. J. Biotechnol. 2010, 9. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.; Duarte, G.; Spohr, M.; Lopes, S.; Fett, J. Distinct physiological responses of two rice cultivars subjected to iron toxicity under field conditions. Ann. Appl. Biol. 2009, 154, 269–277. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R.; Pantoja, O. Progress and challenges for abiotic stress proteomics of crop plants. Proteomics 2013, 13, 1801–1815. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Do Amaral, M.N.; Arge, L.W.P.; Benitez, L.C.; Danielowski, R.; da Silveira Silveira, S.F.; da Rosa Farias, D.; de Oliveira, A.C.; da Maia, L.C.; Braga, E.J.B. Comparative transcriptomics of rice plants under cold, iron, and salt stresses. Funct. Integr. Genom. 2016, 16, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yue, R.; Yuan, C.; Liu, J.; Zhang, L.; Sun, T.; Yang, Y.; Tie, S.; Shen, C. Auxin signaling is involved in iron deficiency-induced photosynthetic inhibition and shoot growth defect in rice (Oryza sativa L.). J. Plant Biol. 2015, 58, 391–401. [Google Scholar] [CrossRef]
- Shanker, A.K.; Cervantes, C.; Loza-Tavera, H.; Avudainayagam, S. Chromium toxicity in plants. Environ. Int. 2005, 31, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Gulli, M.; Rampino, P.; Lupotto, E.; Marmiroli, N.; Perrotta, C. The effect of heat stress and cadmium ions on the expression of a small hsp gene in barley and maize. J. Cereal Sci. 2005, 42, 25–31. [Google Scholar] [CrossRef]
- Nahakpam, S.; Shah, K. Expression of key antioxidant enzymes under combined effect of heat and cadmium toxicity in growing rice seedlings. Plant Growth Regul. 2011, 63, 23–35. [Google Scholar] [CrossRef]
- Panda, S.; Choudhury, S. Chromium stress in plants. Braz. J. Plant Physiol. 2005, 17, 95–102. [Google Scholar] [CrossRef]
- Zeng, F.; Wu, X.; Qiu, B.; Wu, F.; Jiang, L.; Zhang, G. Physiological and proteomic alterations in rice (Oryza sativa L.) seedlings under hexavalent chromium stress. Planta 2014, 240, 291–308. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Bashir, K.; Fujimoto, M.; An, G.; Itai, R.N.; Tsutsumi, N.; Nakanishi, H.; Nishizawa, N.K. Rice-specific mitochondrial iron-regulated gene (MIR) plays an important role in iron homeostasis. Mol. Plant 2009, 2, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Ishimaru, Y.; Shimo, H.; Nagasaka, S.; Fujimoto, M.; Takanashi, H.; Tsutsumi, N.; An, G.; Nakanishi, H.; Nishizawa, N.K. The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2011, 2, 322. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Qiu, B.; Ali, S.; Zhang, G. Genotypic differences in nutrient uptake and accumulation in rice under chromium stress. J. Plant Nutr. 2010, 33, 518–528. [Google Scholar] [CrossRef]
- Huang, T.-L.; Huang, L.-Y.; Fu, S.-F.; Trinh, N.-N.; Huang, H.-J. Genomic profiling of rice roots with short-and long-term chromium stress. Plant Mol. Biol. 2014, 86, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-Y.; Wang, X.; Li, R.; Li, W.-Q.; Chen, K.-M. Genome-wide analysis of the NADK gene family in plants. PLoS ONE 2014, 9, e101051. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H.; Meng, J.L. MapDraw: A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan Hered. 2003, 25, 317–321. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Zhang, M.; Liu, B. Identification of a rice metal tolerance protein OsMTP11 as a manganese transporter. PLoS ONE 2017, 12, e0174987. [Google Scholar] [CrossRef] [PubMed]
- Saifullah; Bibi, S.; Waraich, E.A. Effects of lead forms and organic acids on the growth and uptake of lead in hydroponically grown wheat. Commun. Soil Sci. Plant Anal. 2013, 44, 3150–3160. [Google Scholar]
- Goud, P.B.; Kachole, M.S. Antioxidant enzyme changes in neem, pigeonpea and mulberry leaves in two stages of maturity. Plant Signal. Behav. 2012, 7, 1258–1262. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Gao, D.; Luo, S.; Zeng, R.; Yang, J.; Zhu, X. Physiological and cytological mechanisms of silicon-induced resistance in rice against blast disease. Physiol. Plant. 2008, 134, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Vos, C.; Schat, H.; Waal, M.; Vooijs, R.; Ernst, W. Increased resistance to copper-induced damage of the root cell plasmalemma in copper tolerant Silene cucubalus. Physiol. Plant. 1991, 82, 523–528. [Google Scholar] [CrossRef]
Sample Availability: Currently no samples is available. |




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhammad, I.; Jing, X.-Q.; Shalmani, A.; Ali, M.; Yi, S.; Gan, P.-F.; Li, W.-Q.; Liu, W.-T.; Chen, K.-M. Comparative in Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications. Molecules 2018, 23, 1163. https://doi.org/10.3390/molecules23051163
Muhammad I, Jing X-Q, Shalmani A, Ali M, Yi S, Gan P-F, Li W-Q, Liu W-T, Chen K-M. Comparative in Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications. Molecules. 2018; 23(5):1163. https://doi.org/10.3390/molecules23051163
Chicago/Turabian StyleMuhammad, Izhar, Xiu-Qing Jing, Abdullah Shalmani, Muhammad Ali, Shi Yi, Peng-Fei Gan, Wen-Qiang Li, Wen-Ting Liu, and Kun-Ming Chen. 2018. "Comparative in Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications" Molecules 23, no. 5: 1163. https://doi.org/10.3390/molecules23051163