The Complete Chloroplast Genome of Heimia myrtifolia and Comparative Analysis within Myrtales
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chloroplast Genome Structure and Content
2.2. Codon Usage
2.3. Comparative Genomic Analysis of the cp Genomes in Myrtales
2.3.1. Genome Size Differences between the 12 Myrtales cp Genomes
2.3.2. Contraction and Expansion of All Inverted Repeats (IRs)
2.3.3. Long Repeat Structure Analysis
2.3.4. Simple Sequence Repeat (SSR) Analysis
2.3.5. Divergence Hotspots among Myrtales Species
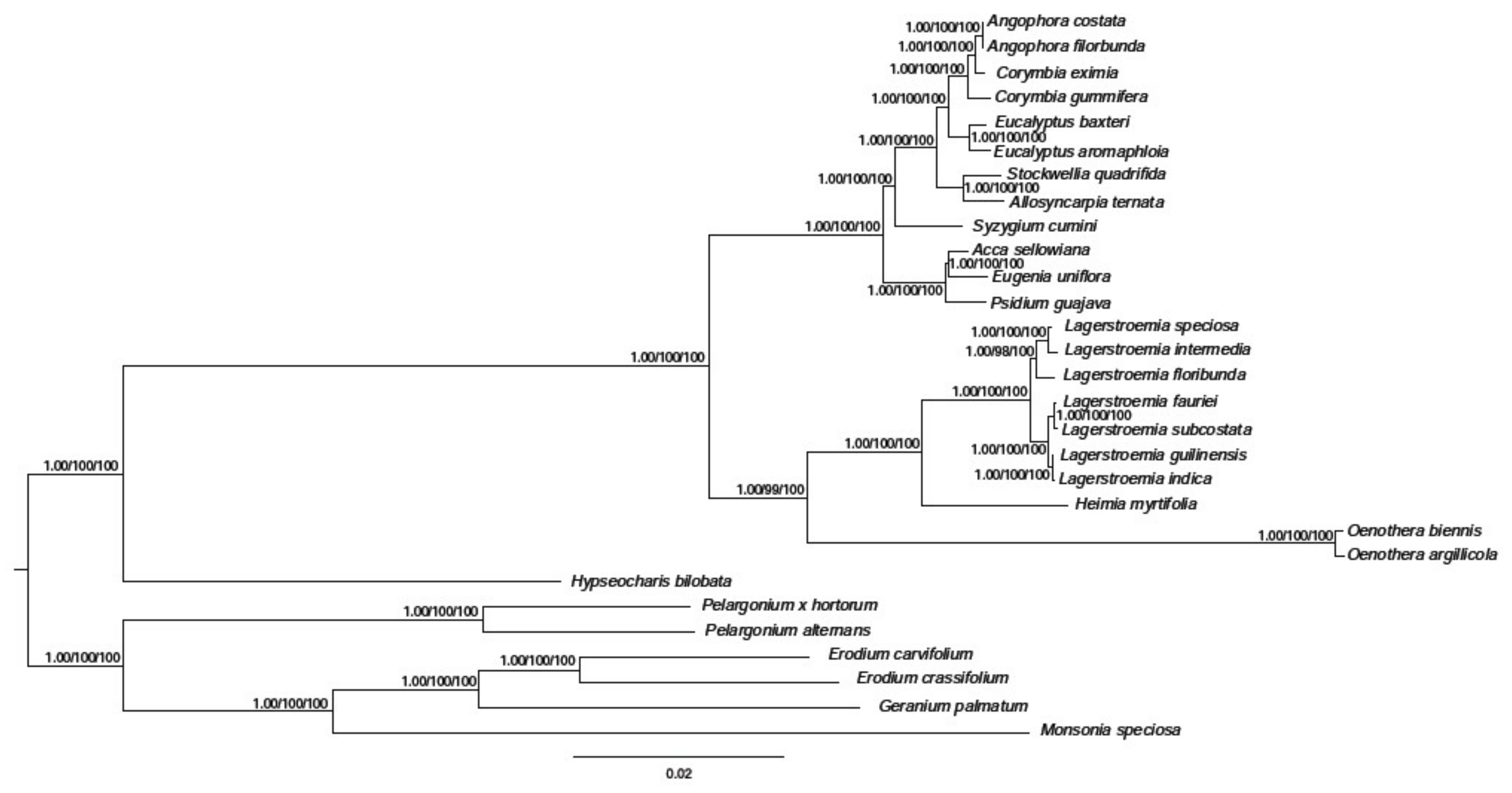
2.3.6. Phylogenetic Analysis of H. myrtifolia and Related Myrtales cp Genomes
3. Materials and Methods
3.1. DNA Extraction of Plant Materials and Sequencing
3.2. Chloroplast Genome Assembly, Annotation, and Structure
3.3. Codon Usage
3.4. Genome Comparative Analysis and Molecular Marker Identification
3.5. IR Expansion and Contraction of cp Boundaries
3.6. Identification of Long Repetitive Sequences and Simple Sequence Repeats (SSRs)
3.7. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gledhill, D. The Names of Plants; Cambridge University Press: Oxford, UK, 1989. [Google Scholar]
- Hegnauer, R.; Herfst, A. Over Heimia salicfolia Link et Otto. Pharm. Weekbl. 1958, 93, 849–865. [Google Scholar] [PubMed]
- Malone, M.H.; Rother, A. Heimia salicifolia: A phytochemical and phytopharmacologic review. J. Ethnopharmacol. 1994, 42, 135–159. [Google Scholar] [CrossRef]
- Lema, W.L.; Blankenship, J.W.; Malone, M.H. Prostaglandin synthetase inhibition by alkaloids of Heimia salicifolia. J. Ethnopharmacol. 1986, 15, 161–167. [Google Scholar] [CrossRef]
- Liu, J.; Qi, Z.; Zhao, Y.; Fu, C.; Xiang, Q.J. Molecular phylogenetics and evolution complete cpDNA genome sequence of Smilax china and phylogenetic placement of Liliales–Influences of gene partitions and taxon sampling. Mol. Phylogenet. Evol. 2012, 64, 545–562. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; DePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Waqas, M.; Khan, A.L.; Khan, M.A.; Kang, S.M.; Imran, Q.M.; Shahzad, R.; Bilal, S.; Yun, B.W.; Lee, I.J. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front. Plant Sci. 2017, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Z.Q.; Bystriakova, N.; Ansell, S.W.; Xiang, Q.P.; Heinrichs, J.; Scheider, H.; Zhang, X.C. Phylogeography of the Sino-Himalayan Fern Lepisorus clathratus on “The Roof of the World”. PLoS ONE 2011, 6, e25896. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Scheider, H.; Wu, Z.Q.; He, L.J.; Zhang, X.C.; Xiang, Q.P. Indehiscent sporangia enable the accumulation of local fern diversity at the Qinghai-Tibetan Plateau. BMC Evol. Biol. 2012, 12, 158. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q. The completed eight chloroplast genomes of tomato from Solanum genus. Mitochondrial DNA Part A 2016, 27, 4155–4157. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q. The whole chloroplast genome of shrub willows (Salix suchowensis). Mitochondrial DNA Part A 2016, 27, 2153–2154. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.H.; Tembrock, L.R.; Ohnson, N.G.; Simmons, M.P.; Wu, Z.Q. The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 Intron from Lagerstroemia (Lythraceae). PLoS ONE 2016, 11, e0150752. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, S.; Li, F.; Zhang, S.; Zhang, H.; Wang, X.; Sun, R.; Bonnema, G.; Borm, T.J. A phylogenetic analysis of chloroplast genomes elucidates the relationships of the six economically important Brassica species comprising the triangle of U. Front. Plant Sci. 2017, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Xue, Q.; Zhu, S.; Sun, J.; Liu, W.; Ding, X. The complete plastome sequences of four Orchid species: Insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhan, D.F.; Jia, X.; Mei, W.L.; Dai, H.F.; Chen, X.T.; Peng, S.Q. Complete chloroplast genome sequence of Aquilaria sinensis (Lour) Gilg and evolution analysis within the Malvales order. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, T.; Yang, J.; Meng, X.; Zhu, J.; Zhao, G. The complete chloroplast genome of Quercus baronii (Quercus L.). Mitochondrial DNA 2015, 1736, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Chaney, L.; Mangelson, R.; Ramaraj, T.; Jellen, E.N.; Maughan, P.J. The complete chloroplast genome sequences for four Amaranthus species (Amaranthaceae). Appl. Plant Sci. 2016, 4, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Mallo, D.; Posada, D. Multilocus inference of species trees and DNA barcoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q.; Tembrock, L.R.; Ge, S. Are differences in genomic data sets due to true biological variants or errors in genome assembly: An example from two chloroplast genomes. PLoS ONE 2015, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cauz-Santos, L.A.; Munhoz, C.F.; Rodde, N.; Cauet, S.; Santos, A.A.; Penha, H.A.; Dornelas, M.C.; Varani, A.M.; Oliveira, G.C.; Bergès, H.; et al. The chloroplast genome of Passiflora edulis (Passifloraceae) sssembled from long sequence reads: Structural organization and phylogenomic studies in Malpighiales. Front. Plant Sci. 2017, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hao, Z.; Xu, H.; Yang, L.; Liu, G.; Sheng, Y.; Zheng, C.; Zheng, W.; Cheng, T.; Shi, J. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front. Plant Sci. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Lv, S.; Zhang, Y.; Du, X.; Wang, L.; Biradar, S.S.; Tan, X.; Wan, F.; Weining, S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 1981, 151, 389–409. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.Y.; Xu, W.; Xing, T.; Zhao, M.M.; Li, Y.L.; Xia, G.M.; Wang, M.C. Synonymous codon usage bias in the plastid genome is unrelated to gene structure and shows evolutionary heterogeneity. Evol. Bioinform. 2015, 11, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H.; Sun, C.; Li, X.; Li, C.; et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef] [PubMed]
- Redwan, R.M.; Saidin, A.; Kumar, S.V. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015, 15, 294. [Google Scholar] [CrossRef] [PubMed]
- Kasiborski, B.A.; Bennett, M.S.; Linton, E.W. The chloroplast genome of Phacus orbicularis (Euglenophyceae): An initial datum point for the phacaceae. J. Phycol. 2016, 52, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean ginseng (Panax ginseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.S.; Li, P.; Qiu, Y.X. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: Comparative genomic and phylogenetic analyses. Front. Plant Sci. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, Z.; Sablok, G.; Daskalova, E.; Zahmanova, G.; Apostolova, E.; Yahubyan, G.; Baev, V. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Amos, W.; Leggo, J.; Goodburn, S.; Jain, S.; Li, S.H.; Margolis, R.L.; Ross, C.A.; Ferguson-Smith, M.A. Microsatellite evolution—Evidence for directionality and variation in rate between species. Nat. Genet. 1995, 10, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Gemayel, R.; Cho, J.; Boeynaems, S.; Verstrepen, K.J. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes 2012, 3, 461–480. [Google Scholar] [CrossRef] [PubMed]
- Voronova, A.; Belevich, V.; Jansons, A.; Rungis, D. Stress-induced transcriptional activation of retrotransposon-like sequences in the Scots pine (Pinus sylvestris L.) genome. Tree Genet. Genomes 2014, 10, 937–951. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, L.; Liu, A.; Chen, J.; Wu, L.; Hu, W.; Zhang, W.; Lee, S.C.; Yang, T.J.; Wang, Y. The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016, 7, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Tripp, E.A. The draft genome of Ruellia speciosa (Beautiful Wild Petunia: Acanthaceae). DNA Res. 2017, 24, 179–192. [Google Scholar] [PubMed]
- Gu, C.H.; Tembrock, L.R.; Zheng, S.Y.; Wu, Z.Q. The complete chloroplast genome of Catha edulis: A comparative analysis of genome features with related species. Int. J. Mol. Sci. 2018, 19, 525. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 33, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wan, J.M. SSRHunter: Development of a local searching software for SSR sites. Yi Chuan 2005, 27, 808–810. [Google Scholar] [PubMed]
- Simmons, M.P.; Cappa, J.J.; Archer, R.H.; Ford, A.J.; Eichstedt, D.; Clevinger, C.C. Phylogeny of the Celastreae (Celastraceae) and the relationships of Catha edulis (qat) inferred from morphological characters and nuclear and plastid genes. Mol. Phylogenet. Evol. 2008, 48, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M. New algorithms and methods to estimate maximum-likelihoods phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. Paup*: Phylogenetic analysis using parsimony (and other methods); Version 4.0b10; 2002; pp. 1–142. [Google Scholar]
Sample Availability: Sample of Heimia myrtifolia is available from the authors. |
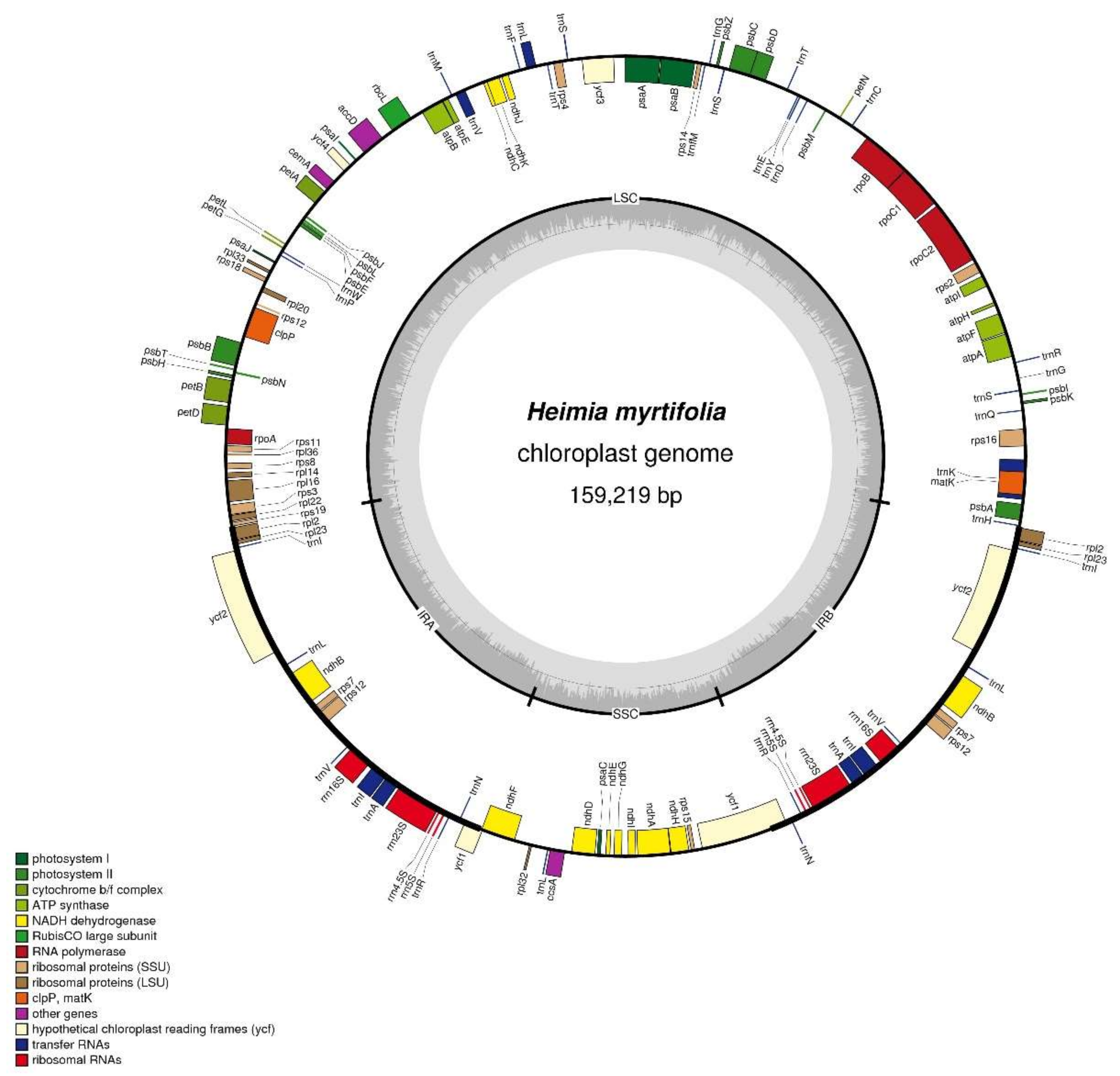
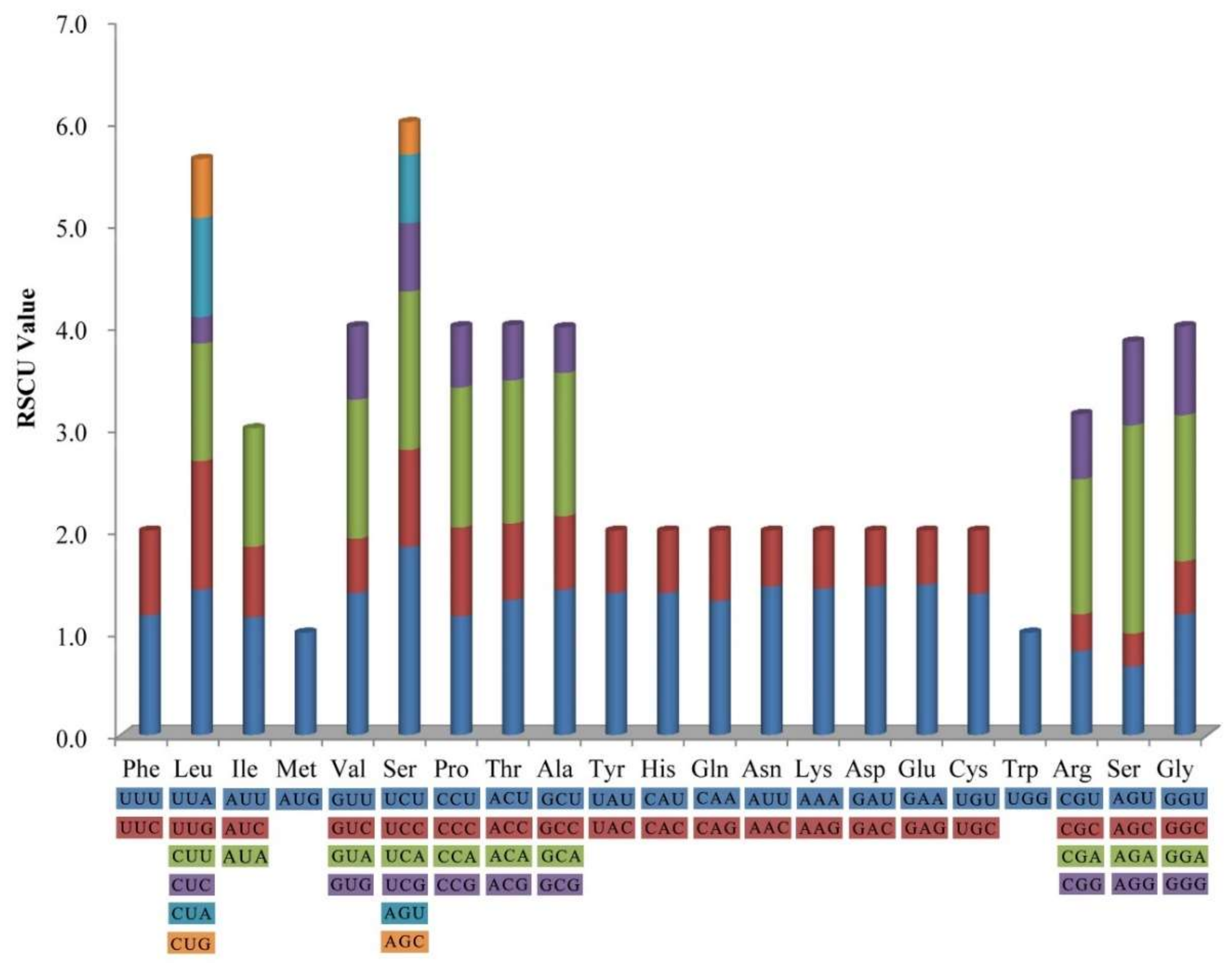
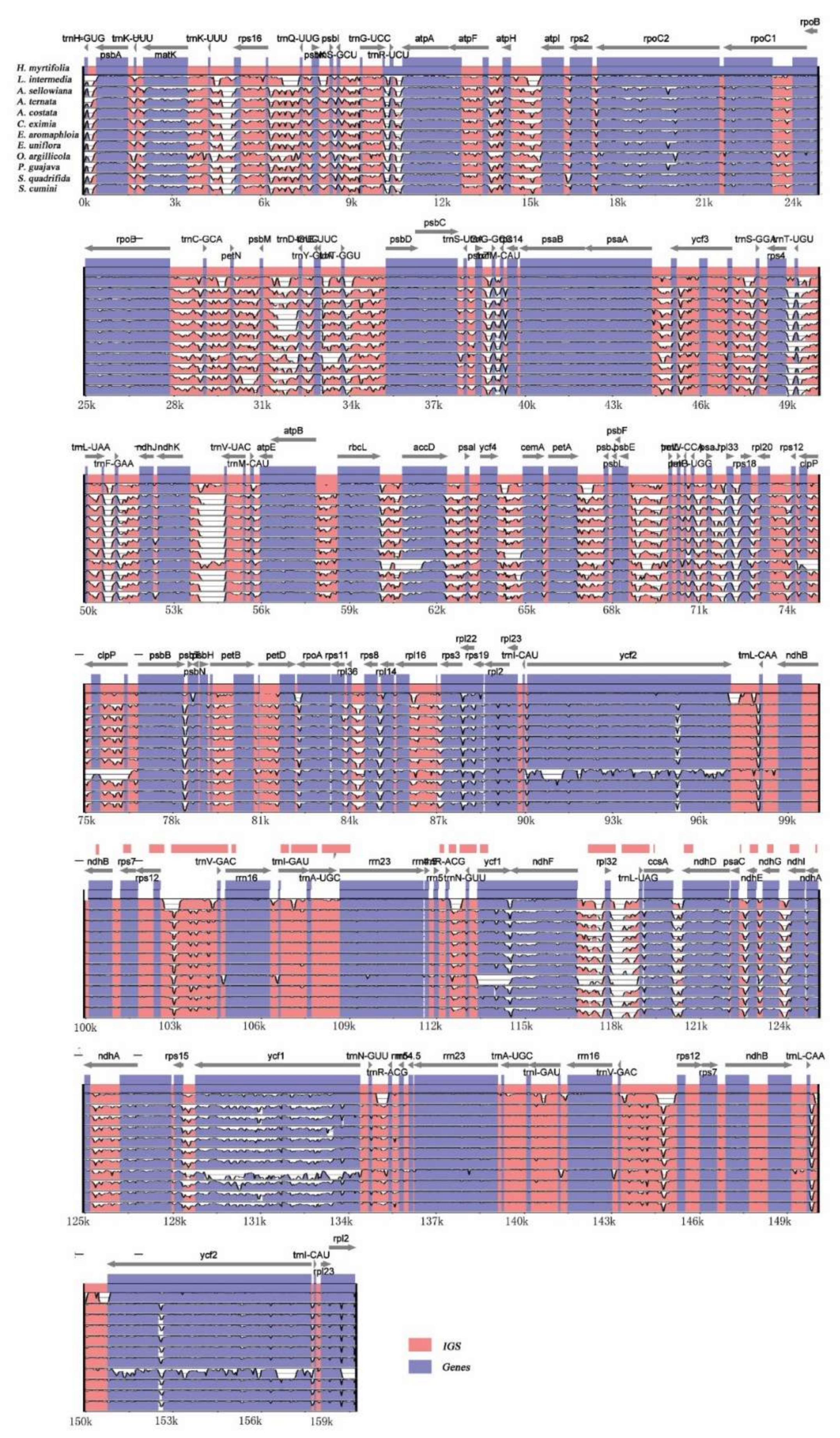
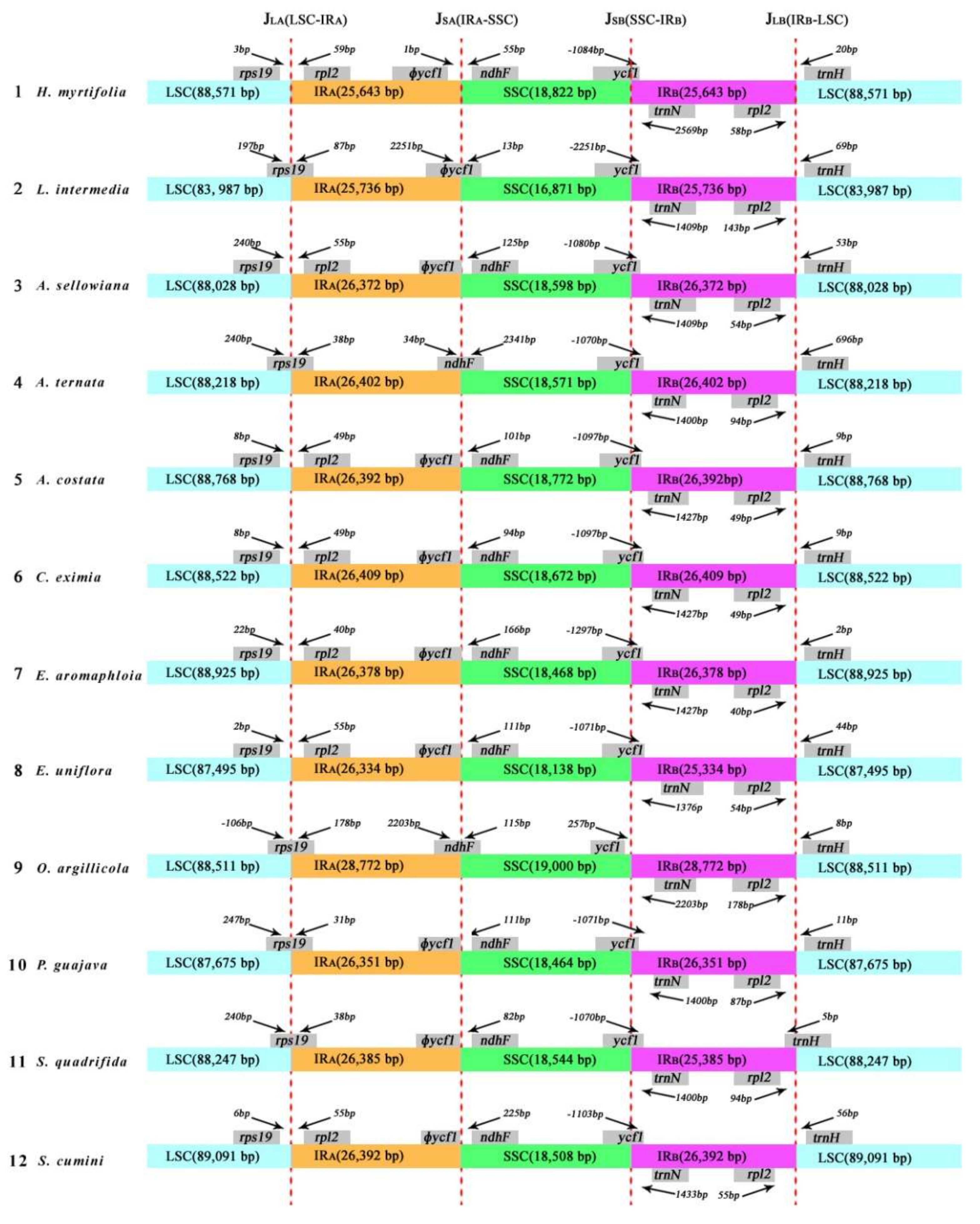

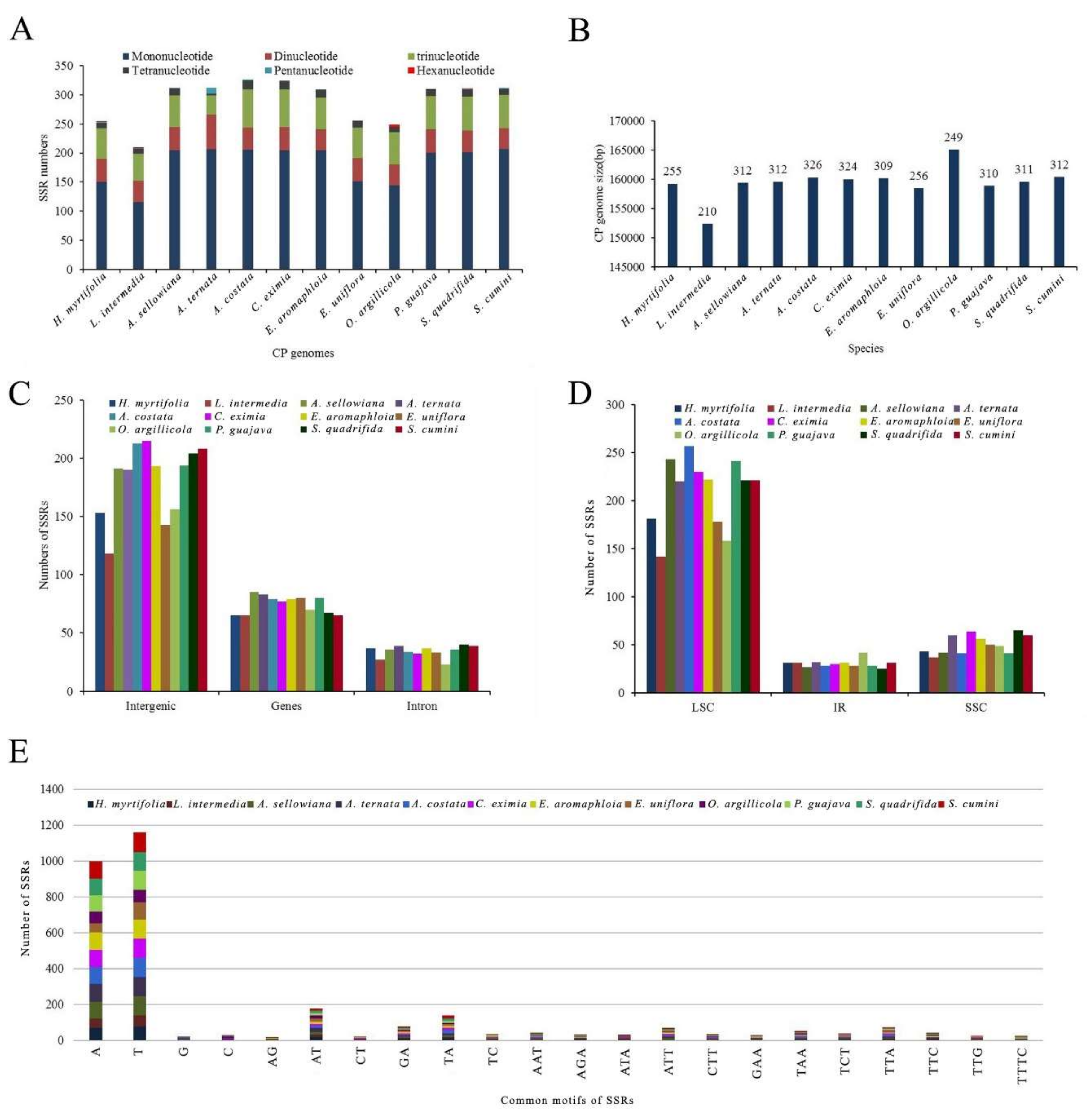

| H. myrtifolia | L. intermedia | A. sellowiana | A. ternata | A. costata | C. eximia | E. aromaphloia | E. uniflora | O. argillicola | P. guajava | S. quadrifida | S. cumini | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession Number | MG921615 | NC034662 | KX289887 | KC180806 | NC022412 | NC022409 | NC022396 | NC027744 | EU262887 | NC033355 | NC022414 | GQ870669 |
| Family | Lythraceae | Lythraceae | Myrtaceae | Myrtaceae | Myrtaceae | Myrtaceae | Myrtaceae | Myrtaceae | Onagraceae | Myrtaceae | Myrtaceae | Myrtaceae |
| Total length (bp) | 159,219 | 152,330 | 159,370 | 159,593 | 160,326 | 160,012 | 160,149 | 158,445 | 165,055 | 158,841 | 159,561 | 160,373 |
| guanine-cytosine (GC) (%) | 37.0 | 37.6 | 37.0 | 37.0 | 37.0 | 37.0 | 37.0 | 37.0 | 37.0 | 39.1 | 37.0 | 37.0 |
| LSC | ||||||||||||
| Length (bp) | 88,571 | 83,987 | 88,028 | 88,218 | 88,768 | 88,522 | 88,925 | 87,459 | 88,511 | 87,675 | 88,247 | 89,091 |
| GC (%) | 35.0 | 36.0 | 35.0 | 35.0 | 35.0 | 35.0 | 35.0 | 35.0 | 37.0 | 35.0 | 35.0 | 35.0 |
| Length (%) | 55.6 | 55.1 | 55.2 | 55.3 | 55.4 | 55.3 | 55.5 | 55.2 | 53.6 | 55.2 | 55.3 | 55.6 |
| SSC | ||||||||||||
| Length (bp) | 18,822 | 16,871 | 18,598 | 18,571 | 18,772 | 18,672 | 18,468 | 18,138 | 19,000 | 18,464 | 18,544 | 18,508 |
| GC (%) | 30.6 | 30.9 | 31.0 | 31.0 | 30.0 | 31.0 | 31.0 | 31.0 | 35.0 | 31.0 | 31.0 | 31.0 |
| Length (%) | 11.8 | 11.1 | 11.7 | 11.6 | 11.7 | 11.7 | 11.5 | 11.4 | 12.0 | 12.0 | 12.0 | 12.0 |
| IRs | ||||||||||||
| Length (bp) | 25,643 | 25,736 | 26,372 | 26,402 | 26,392 | 26,409 | 26,378 | 26,334 | 28,772 | 26,351 | 26,385 | 26,392 |
| GC (%) | 42.6 | 42.5 | 43.0 | 43.0 | 43.0 | 43.0 | 43.0 | 43.0 | 43.0 | 43.0 | 43.0 | 43.0 |
| Length (%) | 16.1 | 16.9 | 16.5 | 16.5 | 16.5 | 16.5 | 16.5 | 16.6 | 35.0 | 35.0 | 33.0 | 33.0 |
| H. myrtifolia | L. intermedia | A. sellowiana | A. ternata | A. costata | C. eximia | E. aromaphloia | E. uniflora | O. argillicola | P. guajava | S. quadrifida | S.cumini | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession Number | MG921615 | NC034662 | KX289887 | KC180806 | NC022412 | NC022409 | NC022396 | NC027744 | EU262887 | NC033355 | NC022414 | GQ870669 |
| Family | Lythraceae | Lythraceae | Myrtaceae | Myrtaceae | Myrtaceae | Myrtaceae | Myrtaceae | Myrtaceae | Onagraceae | Myrtaceae | Myrtaceae | Myrtaceae |
| Protein Coding Genes | ||||||||||||
| Length (bp) | 81,047 | 78,749 | 78,576 | 78,693 | 68,257 | 68,889 | 68,085 | 78,777 | 70,706 | 78,410 | 68,746 | 68,448 |
| GC (%) | 37.0 | 43.0 | 38.0 | 38.0 | 43.0 | 43.0 | 43.0 | 38.0 | 43.0 | 38.0 | 43.0 | 43.0 |
| Length (%) | 51.0 | 52.0 | 49.0 | 49.0 | 43.0 | 43.0 | 43.0 | 50.0 | 43.0 | 49.0 | 43.0 | 43.0 |
| rRNA | ||||||||||||
| Length (bp) | 9050 | 9050 | 9060 | 9056 | 9020 | 9056 | 9056 | 9050 | 9102 | 9056 | 9056 | 9050 |
| GC (%) | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 | 55.0 |
| Length (%) | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 |
| tRNA | ||||||||||||
| Length (bp) | 2817 | 2813 | 2779 | 2716 | 2184 | 2199 | 2270 | 2792 | 2303 | 2790 | 2387 | 2310 |
| GC (%) | 53.0 | 53.0 | 52.0 | 52.0 | 49.0 | 53.0 | 53.0 | 52.0 | 53.0 | 52.0 | 52.0 | 53.0 |
| Length (%) | 2.0 | 2.0 | 2.0 | 2.0 | 1.0 | 1.0 | 1.0 | 2.0 | 1.0 | 2.0 | 1.0 | 1.0 |
| Intergenic Regions | ||||||||||||
| Length (bp) | 50,172 | 46,156 | 51,541 | 51,503 | 65,351 | 64,369 | 65,018 | 49,679 | 69,633 | 50,496 | 63,907 | 65,069 |
| GC (%) | 32.0 | 33.0 | 35.0 | 35.0 | 35.0 | 35.0 | 35.0 | 35.0 | 37.0 | 35.0 | 35.0 | 35.0 |
| Length (%) | 32.0 | 30.0 | 32.0 | 32.0 | 41.0 | 40.0 | 41.0 | 31.0 | 42.0 | 32.0 | 40.0 | 41.0 |
| Intron | ||||||||||||
| Length (bp) | 16,133 | 15,562 | 17,414 | 17,625 | 15,514 | 15,499 | 14,720 | 18,147 | 13,311 | 18,089 | 15,465 | 15,496 |
| GC (%) | 38.0 | 37.0 | 37.0 | 37.0 | 35.0 | 36.0 | 36.0 | 37.0 | 38.0 | 38.0 | 36.0 | 36.0 |
| Length (%) | 10.0 | 10.0 | 11.0 | 11.0 | 10.0 | 10.0 | 9.0 | 11.0 | 8.0 | 11.0 | 10.0 | 10.0 |
| Category of Genes | Function of Genes | Name of Genes |
|---|---|---|
| Subunits of ATP synthase | Genes for photosynthesis | atpA atpB atpE atpFA atpH atpI |
| Subunit of acetyl-CoA-carboxylase | Other genes | accD |
| c-type cytochrome synthesis gene | Other genes | ccsA |
| Envelop membrane protein | Other genes | cemA |
| ATP-dependent protease subunit p gene | Other genes | clpPA |
| Maturase | Other genes | matK |
| Subunits of NADH dehydrogenase | Genes for photosynthesis | ndhAA ndhBA,B ndhC ndhD ndhE ndhF ndhG ndhH ndhI ndhJ ndhK |
| Subunits of photosystem I | Genes for photosynthesis | psaA psaB psaC psaI psaJ |
| Subunits of photosystem II | Genes for photosynthesis | psbA psbB psbC psbD psbE psbF psbH psbI psbJ psbK psbL psbM psbN psbT psbZ |
| Subunits of cytochrome | Genes for photosynthesis | petA petBA petDA petG petL petN |
| Large subunit of Rubisco | Genes for photosynthesis | rbcL |
| Large subunit of ribosome | Self-replication | rpl2B rpl14 rpl16A rpl20 rpl22 rpl23B rpl32 rpl33 rpl36 |
| DNA dependent RNA polymerase | Self-replication | rpoA rpoB rpoC1A rpoC2 |
| Ribosomal RNA genes | Self-replication | rrn16B rrn23B rrn4.5B rrn5B |
| Small subunit of ribosome | Self-replication | rps2 rps3 rps4 rps7B rps8 rps11 rps12A,B rps14 rps15 rps16A rps18 rps19 |
| Transfer RNA genes | Self-replication | trnA-UGCA,B trnC-GCA trnD-GUC trnE-UUC trnF-GAA trnfM-CAU trnG-UCC trnG-GCC trnH-GUG trnI-CAUB trnI-GAUA,B trnK-UUUA trnL-CAAB trnL-UAAA trnL-UAG trnM-CAU trnN-GUUB trnP-UGG trnQ-UUG trnR-ACGB trnR-UCU trnS-GCU trnS-GGA trnS-UGA trnT-GGU trnT-UGU trnV-GACB trnV-UACA trnW-CCA trnY-GUA |
| Conserved open reading frames | Genes of unknown function | ycf1 ycf2B ycf3A ycf4 |
| Gene | Location | ExonI (bp) | IntronI (bp) | ExonII (bp) | IntronII (bp) | ExonIII (bp) |
|---|---|---|---|---|---|---|
| rps16 | LSC | 224 | 861 | 40 | ||
| rpoC1 | LSC | 453 | 743 | 1608 | ||
| atpF | LSC | 145 | 767 | 410 | ||
| petB | LSC | 6 | 780 | 642 | ||
| petD | LSC | 8 | 749 | 475 | ||
| ndhB | IR | 756 | 685 | 777 | ||
| ndhA | SSC | 540 | 1039 | 552 | ||
| rpl16 | LSC | 399 | 976 | 9 | ||
| rps12* | LSC | 114 | 27 | 548 | 231 | |
| ycf3 | LSC | 153 | 796 | 230 | 756 | 124 |
| clpP | LSC | 228 | 585 | 292 | 836 | 71 |
| trnK-UUU | LSC | 35 | 2500 | 37 | ||
| trnL-UAA | LSC | 37 | 532 | 50 | ||
| trnV-UAC | LSC | 37 | 599 | 38 | ||
| trnI-GAU | IR | 35 | 945 | 42 | ||
| trnA-UGC | IR | 35 | 805 | 38 | ||
| trnG-UCC | LSC | 23 | 727 | 52 |
| T | C | A | G | Length (bp) | |
|---|---|---|---|---|---|
| Genome | 31.9 | 18.8 | 31.1 | 18.2 | 159,219 |
| LSC | 33.2 | 17.9 | 31.8 | 17.1 | 88,571 |
| SSC | 34.6 | 16.2 | 34.9 | 14.4 | 18,822 |
| IR | 28.6 | 20.4 | 28.8 | 22.2 | 25,913 |
| tRNA | 22.8 | 26.8 | 23.9 | 26.6 | 2817 |
| rRNA | 19.9 | 25.1 | 24.9 | 30.1 | 9050 |
| Protein-coding genes | 32.1 | 19.4 | 30.2 | 18.4 | 81,047 |
| 1st position | 31.5 | 18.7 | 34.6 | 17.6 | 27,010 |
| 2nd position | 31.2 | 18.7 | 30.7 | 19.4 | 27,010 |
| 3rd position | 33.5 | 23 | 25.2 | 18.2 | 27,010 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, C.; Dong, B.; Xu, L.; Tembrock, L.R.; Zheng, S.; Wu, Z. The Complete Chloroplast Genome of Heimia myrtifolia and Comparative Analysis within Myrtales. Molecules 2018, 23, 846. https://doi.org/10.3390/molecules23040846
Gu C, Dong B, Xu L, Tembrock LR, Zheng S, Wu Z. The Complete Chloroplast Genome of Heimia myrtifolia and Comparative Analysis within Myrtales. Molecules. 2018; 23(4):846. https://doi.org/10.3390/molecules23040846
Chicago/Turabian StyleGu, Cuihua, Bin Dong, Liang Xu, Luke R. Tembrock, Shaoyu Zheng, and Zhiqiang Wu. 2018. "The Complete Chloroplast Genome of Heimia myrtifolia and Comparative Analysis within Myrtales" Molecules 23, no. 4: 846. https://doi.org/10.3390/molecules23040846
APA StyleGu, C., Dong, B., Xu, L., Tembrock, L. R., Zheng, S., & Wu, Z. (2018). The Complete Chloroplast Genome of Heimia myrtifolia and Comparative Analysis within Myrtales. Molecules, 23(4), 846. https://doi.org/10.3390/molecules23040846





