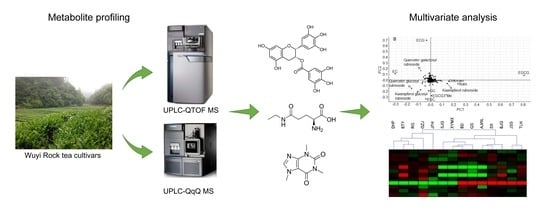
Metabolite Profiling of 14 Wuyi Rock Tea Cultivars Using UPLC-QTOF MS and UPLC-QqQ MS Combined with Chemometrics
Abstract
:1. Introduction
2. Results and Discussion
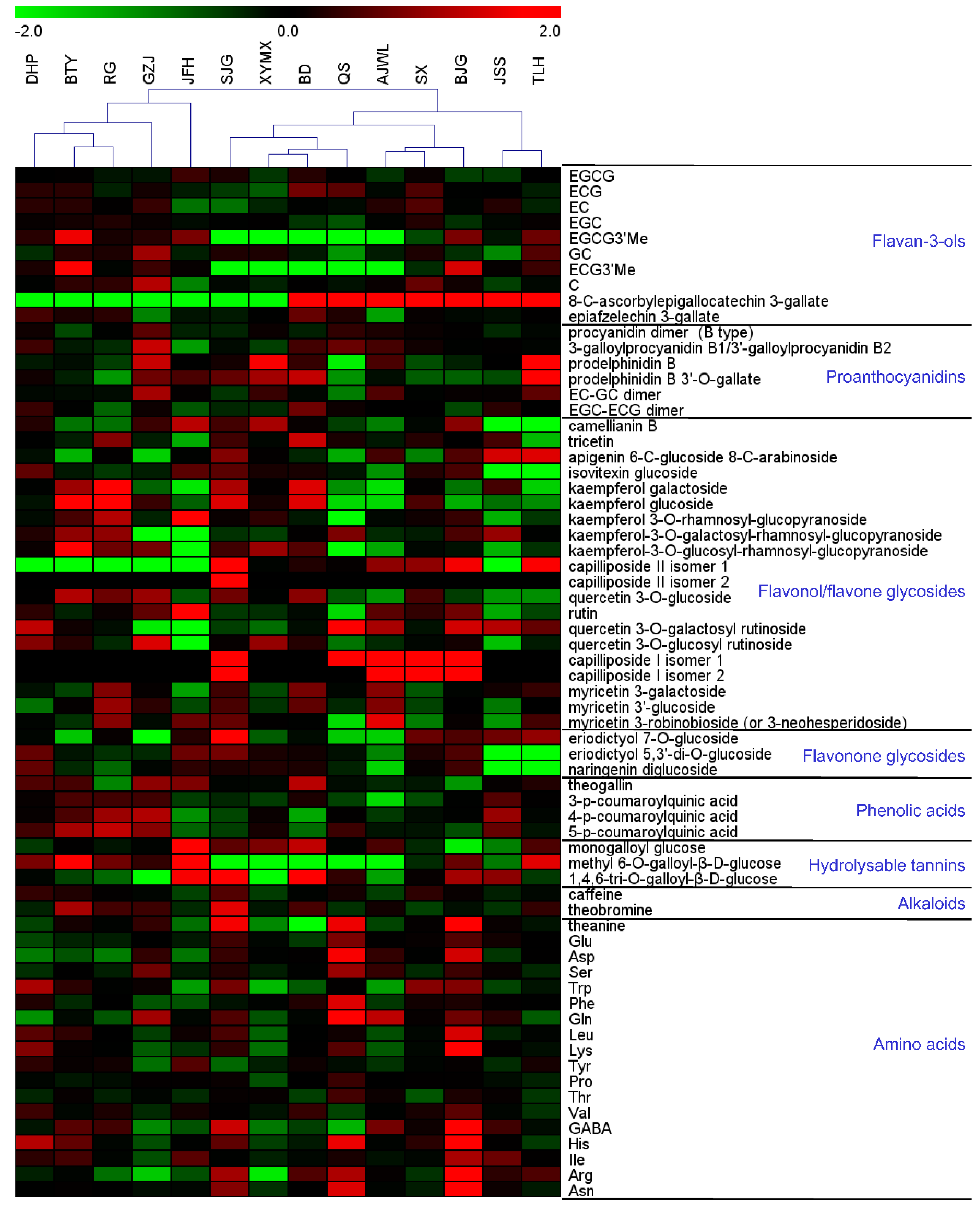
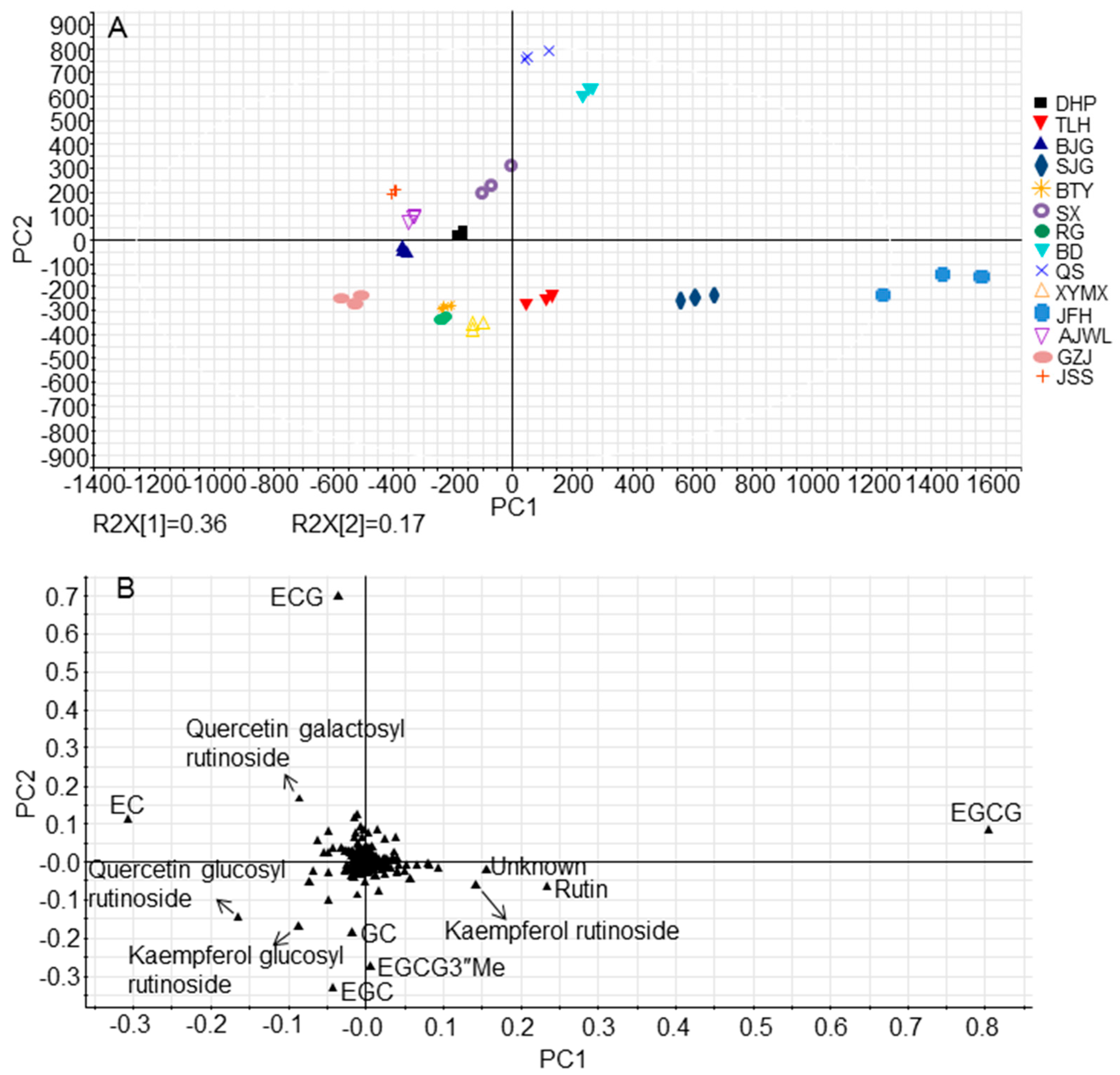
2.1. Major Tea Leaf Metabolites Showed both Universal and Cultivar-Dependent Accumulation Patterns
2.2. Flavan-3-ols Exhibited Variable Levels in Tea Leaves
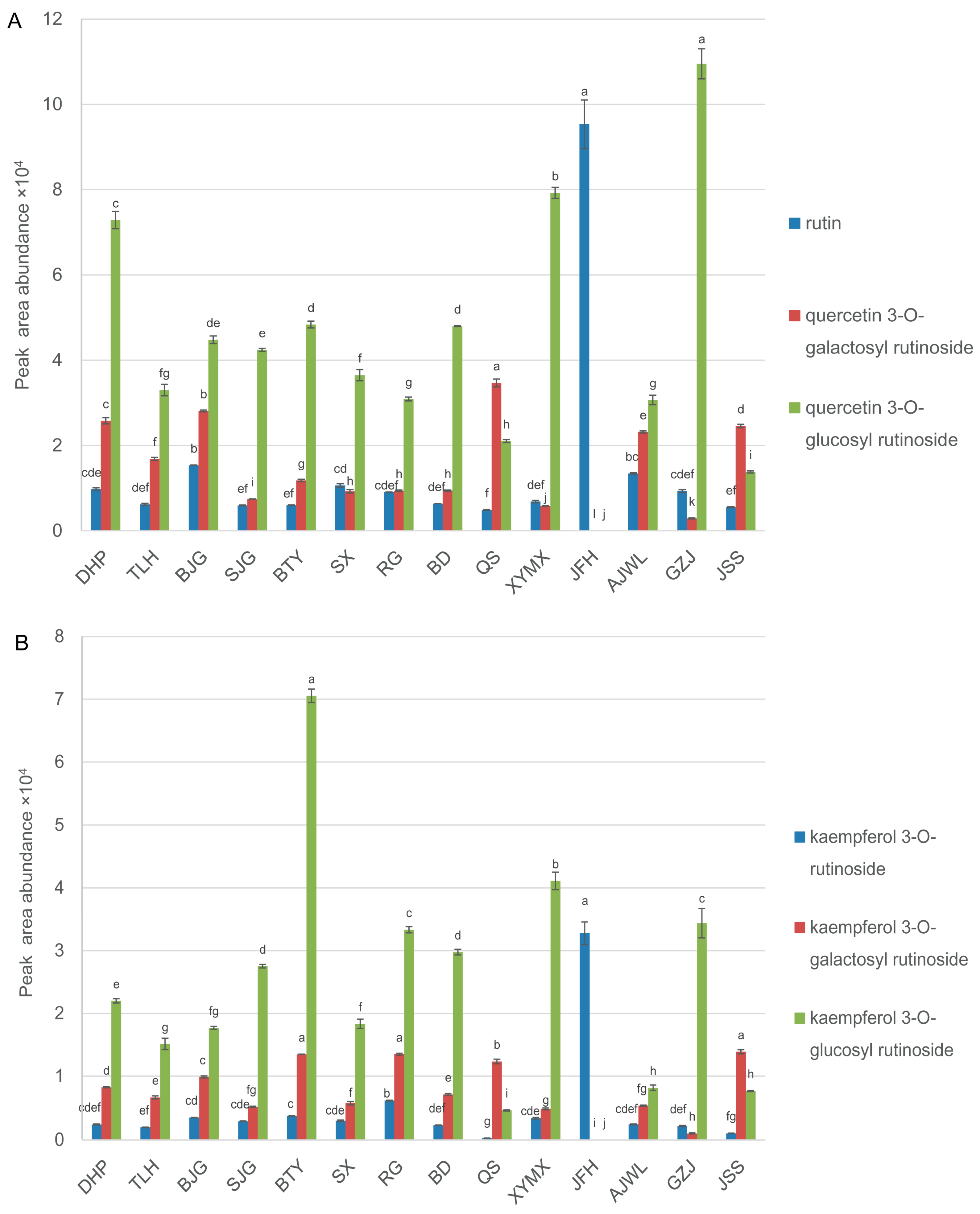
2.3. Cultivar JFH Possessed High Contents of Rutin and Kaempferol Rutinoside
2.4. Cultivars BJG, SJG and QS Demonstrated High Levels of Amino Acids in Leaves
2.5. Purine Alkaloids Exhibited Variable Levels in Tea Leaves
3. Materials and Methods
3.1. Chemicals
3.2. Tea Samples and Sample Preparation
3.3. UPLC-QTOF MS-Based Non-Targeted Metabolite Analysis
3.4. UPLC-QqQ MS-Based Targeted Quantification of Catechins, Rutin, Caffeine and Amino Acids
3.5. Data Processing and Statistical Analysis
3.6. Metabolite Identification
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, Y.L.; Duan, J.; Jiang, Y.M.; Shi, J.; Peng, L.; Xue, S.; Kakuda, Y. Production, quality, and biological effects of Oolong tea (Camellia sinensis). Food Rev. Int. 2011, 27, 1–15. [Google Scholar] [CrossRef]
- Dou, J.; Lee, V.S.; Tzen, J.T.; Lee, M.R. Identification and comparison of phenolic compounds in the preparation of oolong tea manufactured by semifermentation and drying processes. J. Agric. Food Chem. 2007, 55, 7462–7468. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.B. The taste of tea: Material, embodied knowledge and environmental history in northern Fujian, China. J. Mater. Cult. 2017, 22, 3–18. [Google Scholar] [CrossRef]
- Ma, C.H.; Tan, C.; Li, W.L.; Chen, L.B.; Wang, Y.R.; Chen, X. Identification of the different aroma compounds between conventional and freeze dried Wuyi rock Tea (Dangui) using headspace solid phase microextraction. Food Sci. Technol. Res. 2013, 19, 805–811. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. The aroma, taste, color and bioactive constituents of tea. J. Med. Plants Res. 2011, 5, 2110–2124. [Google Scholar]
- Ji, H.G.; Lee, Y.R.; Lee, M.S.; Hwang, K.H.; Kim, E.H.; Park, J.S.; Hong, Y.S. Metabolic phenotyping of various tea (Camellia sinensis L.) cultivars and understanding of their intrinsic metabolism. Food Chem. 2017, 233, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Luo, L.; Li, H.; Liu, R. Phytochemical profiles and antioxidant activity of 27 cultivars of tea. Int. J. Food Sci. Nutr. 2017, 68, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.H.; Cheng, R.H.; Jin, Y.L.; Ding, S.W.; Chen, Z. Evaluation of oolong teas using 1H and 13C solid-state NMR, sensory analysis, and multivariate statistics. J. Chin. Chem. Soc. 2016, 63, 792–799. [Google Scholar] [CrossRef]
- Zhao, F.; Lin, H.T.; Zhang, S.; Lin, Y.F.; Yang, J.F.; Ye, N.X. Simultaneous determination of caffeine and some selected polyphenols in Wuyi Rock tea by high-performance liquid chromatography. J. Agric. Food Chem. 2014, 62, 2772–2781. [Google Scholar] [CrossRef] [PubMed]
- Daglia, M.; Antiochia, R.; Sobolev, A.P.; Mannina, L. Untargeted and targeted methodologies in the study of tea (Camellia sinensis L.). Food Res. Int. 2014, 63, 275–289. [Google Scholar] [CrossRef]
- Dai, W.D.; Qi, D.D.; Yang, T.; Lv, H.P.; Guo, L.; Zhang, Y.; Zhu, Y.; Peng, Q.H.; Xie, D.C.; Tan, J.F.; et al. Nontargeted analysis using ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry uncovers the effects of harvest season on the metabolites and taste quality of tea (Camellia sinensis L.). J. Agric. Food Chem. 2015, 63, 9869–9878. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Q.; Liu, M.; Ma, L.; Shi, Y.; Ruan, J. Metabolomic analyses reveal distinct change of metabolites and quality of green tea during the short duration of a single spring season. J. Agric. Food Chem. 2016, 64, 3302–3309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, Y.; Ma, L.; Yi, X.; Ruan, J. Metabolomic analysis using ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS) uncovers the effects of light intensity and temperature under shading treatments on the metabolites in tea. PLoS ONE 2014, 9, e112572. [Google Scholar] [CrossRef] [PubMed]
- Fraser, K.; Lane, G.A.; Otter, D.E.; Harrison, S.J.; Quek, S.Y.; Hemar, Y.; Rasmussen, S. Non-targeted analysis by LC-MS of major metabolite changes during the oolong tea manufacturing in New Zealand. Food Chem. 2014, 151, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Dai, W.; Lu, M.; Lv, H.; Guo, L.; Zhang, Y.; Zhu, Y.; Peng, Q.; Lin, Z. Study of the dynamic changes in the non-volatile chemical constituents of black tea during fermentation processing by a non-targeted metabolomics approach. Food Res. Int. 2016, 79, 106–113. [Google Scholar] [CrossRef]
- Lee, J.-E.; Lee, B.-J.; Chung, J.-O.; Kim, H.-N.; Kim, E.-H.; Jung, S.; Lee, H.; Lee, S.-J.; Hong, Y.-S. Metabolomic unveiling of a diverse range of green tea (Camellia sinensis) metabolites dependent on geography. Food Chem. 2015, 174, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhang, P.; Pan, Z.; Xu, H.; Luo, Y.; Wang, X. Discrimination of oolong tea (Camellia sinensis) varieties based on feature extraction and selection from aromatic profiles analysed by HS-SPME/GC-MS. Food Chem. 2013, 141, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Alcazar, A.; Ballesteros, O.; Jurado, J.M.; Pablos, F.; Martin, M.J.; Vilches, J.L.; Navalon, A. Differentiation of green, white, black, Oolong, and Pu-erh teas according to their free amino acids content. J. Agric. Food Chem. 2007, 55, 5960–5965. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Gao, M.J.; Hou, R.Y.; Hu, X.Y.; Zhang, L.; Wan, X.C.; Wei, S. Determination of quality constituents in the young leaves of albino tea cultivars. Food Chem. 2014, 155, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Ye, C.X.; Xu, J.K.; Jiang, Y.M. Simultaneous analysis of purine alkaloids and catechins in Camellia sinensis, Camellia ptilophylla and Camellia assamica var. kucha by HPLC. Food Chem. 2007, 100, 1132–1136. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T.; et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, Y.; Wu, Y.; Tan, H.; Meng, F.; Wang, Y.S.; Li, M.; Zhao, L.; Liu, L.; Qian, Y.; et al. Analysis of accumulation patterns and preliminary study on the condensation mechanism of proanthocyanidins in the tea plant [Camellia sinensis]. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Ferreres, F.; Silva, B.M.; Andrade, P.B.; Seabra, R.M.; Ferreira, M.A. Approach to the study of C-glycosyl flavones by ion trap HPLC-PAD-ESI/MS/MS: application to seeds of quince (Cydonia oblonga). Phytochem. Anal. 2003, 14, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Scoparo, C.T.; de Souza, L.M.; Dartora, N.; Sassaki, G.L.; Gorin, P.A.; Iacomini, M. Analysis of Camellia sinensis green and black teas via ultra high performance liquid chromatography assisted by liquid-liquid partition and two-dimensional liquid chromatography (size exclusion x reversed phase). J. Chromatogr. A 2012, 27, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hu, X.; Zhai, Y.; Liu, J.; Wu, G.; Wu, L.; ShenTu, J. Pharmacokinetics and tissue distribution study of camellianin A and its major metabolite in rats by liquid chromatography with tandem mass spectrometry. J. Chromatogr. B 2015, 997, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Xu, L.Z.; Luo, X.Z.; Zhong, Z.; Yang, S.L. Flavonol glycosides from Lysimachia capillipes. J. Asian Nat. Prod. Res. 2002, 4, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Hanhineva, K.; Rogachev, I.; Kokko, H.; Mintz-Oron, S.; Venger, I.; Karenlampi, S.; Aharoni, A. Non-targeted analysis of spatial metabolite composition in strawberry (Fragariaxananassa) flowers. Phytochemistry 2008, 69, 2463–2481. [Google Scholar] [CrossRef] [PubMed]
- Iijima, T.; Mohri, Y.; Hattori, Y.; Kashima, A.; Kamo, T.; Hirota, M.; Kiyota, H.; Makabe, H. Synthesis of (−)-epicatechin 3-(3-O-methylgallate) and (+)-catechin 3-(3-O-methylgallate), and their anti-inflammatory activity. Chem. Biodivers. 2009, 6, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Sano, M.; Suzuki, M.; Miyase, T.; Yoshino, K.; Maeda-Yamamoto, M. Novel antiallergic catechin derivatives isolated from oolong tea. J. Agric. Food Chem. 1999, 47, 1906–1910. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.-P.; Yang, T.; Ma, C.-Y.; Wang, C.-P.; Shi, J.; Zhang, Y.; Peng, Q.-H.; Tan, J.-F.; Guo, L.; Lin, Z. Analysis of naturally occurring 3″-Methyl-epigallocatechin gallate in 71 major tea cultivars grown in China and its processing characteristics. J. Funct. Foods 2014, 7, 727–736. [Google Scholar] [CrossRef]
- Hashimoto, F.; Nonaka, G.; Nishioka, I. Tannins and related-compounds. XC. 8-C-ascorbyl(-)-epigallocatechin 3-O-gallate and novel dimeric flavan-3-ols, oolonghomobisflavans A and B, from oolong tea. Chem. Pharm. Bull. 1989, 37, 3255–3263. [Google Scholar] [CrossRef]
- Hashimoto, F.; Kashiwada, Y.; Nonaka, G.; Nishioka, I.; Nohara, T.; Cosentino, L.M.; Lee, K.H. Evaluation of tea polyphenols as anti-HIV agents. Bioorg. Med. Chem. Lett. 1996, 6, 695–700. [Google Scholar] [CrossRef]
- Nakai, M.; Fukui, Y.; Asami, S.; Toyoda-Ono, Y.; Iwashita, T.; Shibata, H.; Mitsunaga, T.; Hashimoto, F.; Kiso, Y. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J. Agric. Food Chem. 2005, 53, 4593–4598. [Google Scholar] [CrossRef] [PubMed]
- Scharbert, S.; Hofmann, T. Molecular definition of black tea taste by means of quantitative studies, taste reconstitution, and omission experiments. J. Agric. Food Chem. 2005, 53, 5377–5384. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, U.H. Chemistry of Tea. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Braunschweig, Germany, 2013. [Google Scholar]
- Gullon, B.; Lu-Chau, T.A.; Moreira, M.T.; Lema, J.M.; Eibes, G. Rutin: A review on extraction, identification and purification methods, biological activities and approaches to enhance its bioavailability. Trends Food Sci. Technol. 2017, 67, 220–235. [Google Scholar] [CrossRef]
- Ekborg-Ott, K.H.; Taylor, A.; Armstrong, D.W. Varietal differences in the total and enantiomeric composition of theanine in tea. J. Agric. Food Chem. 1997, 45, 353–363. [Google Scholar] [CrossRef]
- Deng, W.-W.; Ogita, S.; Ashihara, H. Biosynthesis of theanine (γ-ethylamino-l-glutamic acid) in seedlings of Camellia sinensis. Phytochem. Lett. 2008, 1, 115–119. [Google Scholar] [CrossRef]
- Deng, W.W.; Ogita, S.; Ashihara, H. Distribution and biosynthesis of theanine in Theaceae plants. Plant Physiol. Biochem. 2010, 48, 70–72. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.-W.; Ashihara, H. Occurrence and de novo biosynthesis of caffeine and theanine in seedlings of tea (Camellia sinensis). Nat. Prod. Commun. 2015, 10, 703–706. [Google Scholar] [PubMed]
- Wu, Q.; Chen, Z.; Sun, W.; Deng, T.; Chen, M. De novo sequencing of the leaf transcriptome reveals complex light-responsive regulatory networks in Camellia sinensis cv. Baijiguan. Front. Plant Sci. 2016, 7, 322. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Xu, Y.X.; Ma, J.Q.; Jin, J.Q.; Huang, D.J.; Yao, M.Z.; Ma, C.L.; Chen, L. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Biol. 2016, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Kitao, N.; Ishida, M.; Morimoto, H.; Irino, F.; Mizuno, K. Expression for caffeine biosynthesis and related enzymes in Camellia sinensis. Z. Naturforsch. C 2010, 65, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.-H.; Zhang, H.-B.; Sheng, J.; Li, K.; Zhang, Q.-J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Wang, L.-Y.; Zhou, J.; He, W.; Zeng, J.-M.; Jiang, Y.-W.; Cheng, H. Comparison of catechins and purine alkaloids in albino and normal green tea cultivars (Camellia sinensis L.) by HPLC. Food Chem. 2012, 130, 720–724. [Google Scholar] [CrossRef]
- Li, C.F.; Yao, M.Z.; Ma, C.L.; Ma, J.Q.; Jin, J.Q.; Chen, L. Differential metabolic profiles during the albescent stages of ‘Anji Baicha’ (Camellia sinensis). PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Y.; Engelhardt, U.H.; Thrane, C.; Maiwald, B.; Stark, J. Determination of flavonol glycosides in green tea, oolong tea and black tea by UHPLC compared to HPLC. Food Chem. 2015, 183, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Kuo, P.-C.; Yang, M.-L.; Li, F.-Y.; Tzen, J.T.C. Effects of baking and aging on the changes of phenolic and volatile compounds in the preparation of old Tieguanyin oolong teas. Food Res. Int. 2013, 53, 732–743. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available. |
| Compound# | Tentative Assignments | RT (min) | Detected [M − H]− (m/z) | Theoretical [M − H]− (m/z) | Mass Error (ppm) | Formula | MS/MS Fragments | Ref. |
|---|---|---|---|---|---|---|---|---|
| Catechins | ||||||||
| 1 | GC | 3.84 | 305.0668 | 305.0661 | 0.51 | C15H14O7 | 219.0660, 179.0348, 167.0347, 139.0397, 125.0242 | Authentic standard b |
| 2 | EGC | 4.93 | 305.0680 | 305.0661 | 1.04 | C15H14O7 | 219.0663, 179.0350, 167.0349, 139.0400, 125.0245 | Authentic standard b |
| 3 | C | 5.36 | 289.0718 | 289.0712 | 0.20 | C15H14O6 | 245.0817, 203.0710, 125.0242 | Authentic standard b |
| 4 | EC | 6.28 | 289.0722 | 289.0712 | 2.13 | C15H14O6 | 245.0820, 203.0711, 123.0450 | Authentic standard b |
| 5 | EGCG | 6.35 | 457.0783 | 457.0771 | 2.60 | C22H18O11 | 305.0662, 169.0143, 125.0244 | Authentic standard b |
| 6 | 8-C-ascorbylepigallocatechin 3-gallate | 6.65 | 631.0938 | 631.0935 | -0.37 | C28H24O17 | 479.0821, 316.0218 | [21] |
| 7 | EGCG3″Me | 7.42 | 471.0933 | 471.0927 | 0.16 | C23H20O11 | 305.0667, 287.0560, 183.0300, 161.0243 | Authentic standard b |
| 8 | ECG | 7.86 | 441.0827 | 441.0822 | 0.51 | C22H18O10 | 331.0458, 289.0719, 245.0818, 169.0145, 125.0245 | Authentic standard b |
| 9 | ECG3″Me | 8.92 | 455.0984 | 455.0978 | -0.02 | C23H20O10 | 289.0717, 183.0298 | Authentic standard b |
| 10 | epiafzelechin 3-gallate | 8.97 | 425.0880 | 425.0873 | 0.41 | C22H18O9 | 273.0765, 255.0661 | [21] |
| Proanthocyanidins | ||||||||
| 11 | prodelphinidin B | 4.11 | 609.1251 | 609.1244 | 0.13 | C30H26O14 | 441.0825, 423.0718, 305.0667, 125.0243 | [2] |
| 12 | EC-GC dimer | 4.80 | 593.1301 | 593.1295 | 0.06 | C30H26O13 | 423.0714, 305.0659, 289.0717 | [21] |
| 13 | prodelphinidin B2 (or B4) 3′-O-gallate | 5.12 | 761.1357 | 761.1354 | -0.27 | C37H30O18 | 609.1236, 591.1144, 577.1347, 423.0717 | [2] |
| 14 | procyanidin dimer (B type) | 5.68 | 577.1352 | 577.1346 | 0.02 | C30H26O12 | 451.1029, 425.0874, 407.0768, 289.0716 | [22] |
| 15 | EGC-ECG dimer | 6.04 | 745.1409 | 745.1405 | -0.19 | C37H30O17 | 593.1298, 423.0714, 407.0768, 177.0191 | [23] |
| 16 | 3-galloylprocyanidin B1/3′-galloylprocyanidin B2 | 6.78 | 729.1458 | 729.1456 | -0.36 | C37H30O16 | 407.0766, 289.0716 | [23] |
| Flavonol/flavone glycosides | ||||||||
| 17 | isovitexin glucoside | 6.08 | 595.1655 a | 595.1663 a | -0.40 | C27H30O15 | 313.0711 | [24] |
| 18 | apigenin 6-C-glucoside 8-C-arabinoside | 6.91 | 563.1405 | 563.1401 | -0.22 | C26H28O14 | 473.1086, 443.0980, 383.0769, 353.0664 | [24] |
| 19 | myricetin 3-robinobioside (or 3-neohesperidoside) | 6.93 | 625.1407 | 625.1405 | -0.44 | C27H30O17 | 316.0219 | [21] |
| 20 | myricetin 3-galactoside | 7.02 | 479.0829 | 479.0826 | 0.36 | C21H20O13 | 316.0223, 315.0141, 271.0245 | [25] |
| 21 | myricetin 3′-glucoside | 7.12 | 479.0830 | 479.0826 | -0.22 | C21H20O13 | 316.0224, 315.0146, 271.0245 | [25] |
| 22 | quercetin 3-O-galactosyl rutinoside | 7.21 | 771.1990 | 771.1984 | 0.03 | C33H40O21 | 609.1434, 463.0903, 301.0339, 300.0266 | [11] |
| 23 | quercetin 3-O-glucosyl rutinoside | 7.36 | 771.1991 | 771.1984 | 0.23 | C33H40O21 | 609.1458, 301.0348, 300.0272 | [11] |
| 24 | camellianin B | 7.69 | 579.1704 a | 579.1714 a | -0.75 | C27H30O14 | 433.1129, 313.0709 | [26] |
| 25 | rutin | 7.70 | 609.1455 | 609.1456 | -0.93 | C27H30O16 | 300.0274, 299.0195 | Authentic standard b |
| 26 | kaempferol-3-O-galactosylrutinoside | 7.72 | 755.2040 | 755.2035 | -0.05 | C33H40O20 | 533.1294, 285.0398, 284.0319 | [2] |
| 27 | tricetin | 7.90 | 303.0504a | 303.0505 a | 1.56 | C15H10O7 | 285.0402, 257.0450 | [21] |
| 28 | kaempferol-3-O-glucosylrutinoside | 8.00 | 755.2042 | 755.2035 | 0.19 | C33H40O20 | 593.1511, 285.0403, 284.0325 | [2] |
| 29 | quercetin 3-O-glucoside | 8.02 | 463.0879 | 463.0877 | -0.62 | C21H20O12 | 300.0274, 299.0195, 243.0297 | [25] |
| 30 | kaempferol 3-O-rutinoside | 8.43 | 593.1511 | 593.1506 | -0.21 | C27H30O15 | 501.0102, 285.0399, 284.0326 | Authentic standard b |
| 31 | kaempferol galactoside | 8.51 | 447.0929 | 447.0927 | -0.84 | C21H20O11 | 285.0387, 284.0317 | |
| 32 | kaempferol glucoside | 8.78 | 447.0929 | 447.0927 | -0.76 | C21H20O11 | 284.0324, 255.0295, 227.0349 | Authentic standard b |
| 33 | capilliposide I isomer 1 | 9.94 | 1065.3052 a | 1065.3087 a | -2.68 | C48H56O27 | 617.2078, 449.1078, 303.0506 | [27] |
| 34 | capilliposide II isomer 1 | 10.19 | 1049.3113 a | 1049.3138 a | -1.84 | C48H56O26 | 741.2036, 595.1495, 287.0553 | [27] |
| 35 | capilliposide I isomer 2 | 10.60 | 1065.3061 a | 1065.3087 a | -1.97 | C48H56O27 | 617.2083, 449.1086, 303.0514 | [27] |
| 36 | capilliposide II isomer 2 | 10.88 | 1049.3114 a | 1049.3138 a | -1.78 | C48H56O26 | 741.2048, 287.0564 | [27] |
| Flavonone glycosides | ||||||||
| 37 | eriodictyol 5,3′-di-O-glucoside | 6.08 | 611.1617 | 611.1612 | -0.10 | C27H32O16 | 491.1189, 449.1292, 329.0869 | [21] |
| 38 | naringenin diglucoside | 6.16 | 595.1664 | 595.1663 | -0.78 | C27H32O15 | 577.1552, 475.1243, 433.1348, 381.0827, 313.0923 | [21] |
| 39 | eriodictyol 7-O-glucoside | 6.57 | 449.1086 | 449.1084 | -0.68 | C21H22O11 | 329.0657, 197.0455 | [21] |
| Phenolic acids | ||||||||
| 40 | theogallin | 2.90 | 343.0669 | 343.0665 | 0.16 | C14H16O10 | 191.0560 | Authentic standard b |
| 41 | 3-p-coumaroylquinic acid | 5.18 | 337.0928 | 337.0923 | -0.25 | C16H18O8 | 163.0399 | [2] |
| 42 | 4-p-coumaroylquinic acid | 6.15 | 337.0924 | 337.0923 | -0.36 | C16H18O8 | 191.0542, 173.0454, 163.0398,119.0500,111.0441, 93.0343 | [2] |
| 43 | 5-p-coumaroylquinic acid | 6.42 | 337.0925 | 337.0923 | -0.08 | C16H18O8 | 173.0457, 163.0396, 119.0499, 93.0343 | [2] |
| Hydrolysable tannins | ||||||||
| 44 | monogalloyl glucose | 2.44 | 331.0668 | 331.0665 | -0.80 | C13H16O10 | 271.0454, 211.0247, 169.0140, 125.0242 | [28] |
| 45 | methyl 6-O-galloyl-β-d-glucose | 3.67 | 345.0823 | 345.0822 | -1.09 | C14H18O10 | 285.0611, 225.0401, 183.0296 | [21] |
| 46 | 1,4,6-tri-O-galloyl-β-d-glucose | 6.64 | 635.0894 | 635.0884 | 1.60 | C27H24O18 | 483.0777, 465.0666, 423.0524, 313.0562, 241.0348, 169.0142, 125.0236 | [25] |
| Alkaloids | ||||||||
| 47 | theobromine | 3.80 | 181.0725 a | 181.0726 a | 3.53 | C7H8N4O2 | 138.0671 | Authentic standard b |
| 48 | caffeine | 5.60 | 195.0893 a | 195.0882 a | 5.60 | C8H10N4O2 | 138.0670 | Authentic standard b |
| Amino acids | ||||||||
| 49 | theanine | 1.43 | 175.1085 a | 175.1083 a | 4.72 | C7H14N2O3 | 158.0823, 129.1030 | Authentic standard b |
| Compound | DHP | TLH | BJG | SJG | BTY | SX | RG | BD | QS | XYMX | JFH | AJWL | GZJ | JSS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catechins | 210.09 ± 12.55 bc | 201.42 ± 5.35 c | 155.29 ± 6.42 e | 199.58 ± 4.77 cd | 225.73 ± 6.42 ab | 233.28 ± 4.11 a | 195.99 ± 8.44 cd | 210.14 ± 16.03 abc | 177.60 ± 2.85 de | 162.51 ± 5.88 e | 218.92 ± 3.72 abc | 167.86 ± 4.68 e | 201.94 ± 9.50 c | 165.06 ± 6.09 e |
| EGCG | 94.67 ± 5.17 d | 95.07 ± 2.93 d | 66.93 ± 3.97 f | 111.71 ± 1.76 bc | 97.81 ± 2.50 d | 103.73 ± 1.66 cd | 81.87 ± 3.43 e | 116.21 ± 8.70 b | 96.45 ± 2.08 d | 69.71 ± 2.25 f | 128.08 ± 1.63 a | 71.20 ± 2.23 ef | 81.60 ± 3.61 e | 68.27 ± 3.35 f |
| ECG | 22.83 ± 1.56 c | 15.17 ± 0.56 fg | 18.13 ± 0.93 de | 13.09 ± 0.32 gh | 22.83 ± 0.60 c | 26.51 ± 0.26 b | 14.91 ± 0.67 fg | 31.31 ± 2.56 a | 27.92 ± 0.29 b | 11.12 ± 0.29 h | 15.28 ± 0.14 fg | 16.77 ± 0.32 ef | 20.80 ± 0.89 bc | 18.48 ± 0.71 de |
| EC | 11.73 ± 0.70 b | 7.60 ± 0.14 d | 9.04 ± 0.28 c | 5.12 ± 0.14 e | 11.65 ± 0.58 b | 13.95 ± 0.40 a | 9.73 ± 0.68 c | 9.15 ± 0.74 c | 8.59 ± 0.30 cd | 7.55 ± 0.20 d | 5.09 ± 0.17 e | 11.44 ± 0.21 b | 12.43 ± 0.39 b | 11.41 ± 0.28 b |
| EGC | 72.83 ± 5.50 abcd | 71.60 ± 1.42 bcd | 50.99 ± 1.45 g | 65.09 ± 2.37 def | 76.43 ± 2.91 abc | 81.44 ± 2.20 a | 80.21 ± 3.28 ab | 49.92 ± 3.92 g | 42.48 ± 0.65 g | 69.68 ± 3.09 cde | 60.53 ± 2.29 f | 64.80 ± 1.81 def | 73.84 ± 3.98 abc | 61.44 ± 1.89 ef |
| EGCG3″Me | 4.81 ± 0.22 c | 6.48 ± 0.36 b | 6.89 ± 0.49 b | 0.03 ± 0.00 f | 12.05 ± 0.45 a | 2.62 ± 0.15 e | 4.49 ± 0.05 c | 0.03 ± 0.00 f | 0.05 ± 0.00 f | 0.03 ± 0.00 f | 6.89 ± 0.39 b | 0.13 ± 0.01 f | 4.77 ± 0.18 c | 3.38 ± 0.18 d |
| GC | 2.48 ± 0.14 d | 4.80 ± 0.16 b | 2.45 ± 0.05 d | 3.84 ± 0.21 c | 4.00 ± 0.16 c | 3.81 ± 0.05 c | 3.81 ± 0.33 c | 2.75 ± 0.30 d | 1.44 ± 0.00 e | 3.81 ± 0.09 c | 2.67 ± 0.05 d | 2.69 ± 0.12 d | 6.75 ± 0.47 a | 1.55 ± 0.05 e |
| C | 0.75 ± 0.05 de | 0.69 ± 0.12 def | 0.85 ± 0.05 cd | 0.69 ± 0.05 def | 0.96 ± 0.08 c | 1.23 ± 0.05 b | 0.96 ± 0.08 c | 0.77 ± 0.05 cde | 0.67 ± 0.05 def | 0.61 ± 0.05 ef | 0.37 ± 0.05 g | 0.83 ± 0.05 cd | 1.76 ± 0.08 a | 0.53 ± 0.05 fg |
| Rutin | 0.68 ± 0.02 bcd | 0.38 ± 0.01 de | 0.89 ± 0.12 b | 0.40 ± 0.04 de | 0.44 ± 0.02 de | 0.70 ± 0.07 bcd | 0.60 ± 0.04 bcd | 0.50 ± 0.07 cde | 0.18 ± 0.02 e | 0.42 ± 0.01 de | 5.40 ± 0.39 a | 0.83 ± 0.11 bc | 0.85 ± 0.08 bc | 0.23 ± 0.02 e |
| Caffeine | 26.00 ± 2.28 b | 18.35 ± 0.40 de | 21.81 ± 0.44 cd | 29.79 ± 1.40 a | 23.97 ± 1.09 bc | 25.41 ± 1.11 b | 20.93 ± 1.38 cd | 19.28 ± 1.42 de | 22.93 ± 0.62 bc | 14.27 ± 1.02 f | 13.81 ± 0.58 f | 14.16 ± 0.76 f | 18.85 ± 1.72 de | 16.51 ± 0.36 ef |
| Amino acids | 4.60 ± 0.32 g | 6.01 ± 0.36 ef | 21.07 ± 0.80 a | 15.83 ± 0.72 c | 6.68 ± 0.31 ef | 6.84 ± 0.06 ef | 5.83 ± 0.22 f | 3.11 ± 0.15 h | 18.24 ± 0.41 b | 4.29 ± 0.39 g | 4.33 ± 0.17 g | 5.77 ± 0.35 f | 8.05 ± 0.23 d | 7.09 ± 0.19 de |
| l-Theanine | 2.61 ± 0.30 ef | 3.31 ± 0.33 de | 14.32 ± 0.68 a | 12.08 ± 0.77 b | 4.35 ± 0.36 cd | 3.97 ± 0.05 cd | 3.63 ± 0.28 de | 0.80 ± 0.08 g | 11.63 ± 0.62 b | 1.81 ± 0.23 fg | 2.08 ± 0.08 f | 2.59 ± 0.26 ef | 4.91 ± 0.30 c | 4.13 ± 0.18 cd |
| Glu | 1.15 ± 0.05 g | 1.73 ± 0.10 cde | 2.53 ± 0.10 b | 2.33 ± 0.11 b | 1.38 ± 0.07 fg | 1.82 ± 0.07 cd | 1.41 ± 0.09 fg | 1.22 ± 0.05 g | 3.05 ± 0.16 a | 1.57 ± 0.08 def | 1.45 ± 0.08 efg | 1.76 ± 0.15 cd | 1.68 ± 0.05 cdef | 1.93 ± 0.17 c |
| Asp | 0.22 ± 0.03 e | 0.43 ± 0.01 d | 1.13 ± 0.05 b | 0.56 ± 0.02 c | 0.27 ± 0.01 e | 0.42 ± 0.03 d | 0.22 ± 0.02 e | 0.43 ± 0.02 d | 1.37 ± 0.06 a | 0.41 ± 0.05 d | 0.23 ± 0.02 e | 0.55 ± 0.01 c | 0.55 ± 0.03 c | 0.31 ± 0.04 e |
| Ser | 0.22 ± 0.01 h | 0.23 ± 0.03 gh | 0.39 ± 0.02 c | 0.35 ± 0.04 cde | 0.29 ± 0.02 efg | 0.22 ± 0.02 gh | 0.24 ± 0.01 gh | 0.28 ± 0.01 efgh | 0.58 ± 0.01 a | 0.24 ± 0.04 gh | 0.24 ± 0.02 fgh | 0.37 ± 0.02 cd | 0.48 ± 0.03 b | 0.31 ± 0.02 def |
| Trp | 0.06 ± 0.00 a | 0.02 ± 0.00 def | 0.05 ± 0.00 b | 0.05 ± 0.00 b | 0.03 ± 0.00 c | 0.05 ± 0.00 b | 0.03 ± 0.00 cde | 0.02 ± 0.00 fgh | 0.03 ± 0.00 cd | 0.01 ± 0.00 h | 0.01 ± 0.00 gh | 0.01 ± 0.00 gh | 0.03 ± 0.00 cd | 0.02 ± 0.00 efg |
| Phe | 0.05 ± 0.00 bc | 0.04 ± 0.00 bcde | 0.05 ± 0.00 bc | 0.04 ± 0.00 def | 0.03 ± 0.00 efg | 0.05 ± 0.00 bc | 0.04 ± 0.00 bcde | 0.05 ± 0.00 b | 0.12 ± 0.01 a | 0.04 ± 0.00 cdef | 0.03 ± 0.00 g | 0.03 ± 0.00 fg | 0.03 ± 0.00 g | 0.04 ± 0.00 bcd |
| Gln | 0.05 ± 0.00 g | 0.07 ± 0.01 fg | 0.18 ± 0.01 c | 0.16 ± 0.02 cd | 0.10 ± 0.00 ef | 0.12 ± 0.01 de | 0.07 ± 0.00 fg | 0.10 ± 0.01 ef | 1.19 ± 0.05 a | 0.07 ± 0.01 fg | 0.10 ± 0.00 ef | 0.27 ± 0.01 b | 0.23 ± 0.01 b | 0.14 ± 0.00 de |
| Leu | 0.04 ± 0.00 b | 0.02 ± 0.00 ef | 0.07 ± 0.00 a | 0.04 ± 0.00 bc | 0.03 ± 0.00 cd | 0.02 ± 0.00 efg | 0.03 ± 0.00 ef | 0.03 ± 0.00 ef | 0.03 ± 0.00 de | 0.02 ± 0.00 i | 0.03 ± 0.00 de | 0.02 ± 0.00 hi | 0.02 ± 0.00 ghi | 0.02 ± 0.00 fgh |
| Lys | 0.04 ± 0.00 b | 0.02 ± 0.00 defgh | 0.15 ± 0.01 a | 0.02 ± 0.00 cd | 0.02 ± 0.00 de | 0.02 ± 0.00 defgh | 0.02 ± 0.00 defg | 0.02 ± 0.00 def | 0.03 ± 0.00 bc | 0.01 ± 0.00 h | 0.01 ± 0.00 efgh | 0.01 ± 0.00 fgh | 0.01 ± 0.00 gh | 0.02 ± 0.00 de |
| Tyr | 0.03 ± 0.00 b | 0.02 ± 0.00 bc | 0.02 ± 0.00 cde | 0.01 ± 0.00 hi | 0.02 ± 0.00 bcd | 0.02 ± 0.00 def | 0.02 ± 0.00 bc | 0.02 ± 0.00 bcd | 0.02 ± 0.00 efg | 0.02 ± 0.00 fg | 0.03 ± 0.00 a | 0.02 ± 0.00 gh | 0.01 ± 0.00 i | 0.02 ± 0.00 fg |
| Pro | 0.02 ± 0.00 cde | 0.02 ± 0.00 f | 0.03 ± 0.00 bc | 0.03 ± 0.00 b | 0.02 ± 0.00 ef | 0.03 ± 0.00 bc | 0.02 ± 0.00 def | 0.03 ± 0.00 bcd | 0.03 ± 0.00 a | 0.02 ± 0.00 g | 0.03 ± 0.00 bc | 0.03 ± 0.00 bcd | 0.03 ± 0.00 bc | 0.02 ± 0.00 def |
| Thr | 0.02 ± 0.00 bcde | 0.02 ± 0.00 cde | 0.03 ± 0.00 ab | 0.03 ± 0.01 abcd | 0.03 ± 0.01 abcd | 0.02 ± 0.00 e | 0.02 ± 0.00 bcde | 0.02 ± 0.00 de | 0.04 ± 0.01 a | 0.03 ± 0.00 abcd | 0.02 ± 0.00 cde | 0.03 ± 0.00 abc | 0.03 ± 0.00 bcde | 0.03 ± 0.00 abcd |
| Val | 0.02 ± 0.00 ab | 0.01 ± 0.00 fg | 0.02 ± 0.00 a | 0.02 ± 0.00 cde | 0.01 ± 0.00 defg | 0.02 ± 0.00 bcd | 0.02 ± 0.00 bc | 0.02 ± 0.00 ab | 0.01 ± 0.00 g | 0.01 ± 0.00 efg | 0.02 ± 0.00 bcd | 0.01 ± 0.00 cdef | 0.01 ± 0.00 efg | 0.01 ± 0.00 cdef |
| GABA | 0.02 ± 0.00 fg | 0.02 ± 0.00 ef | 0.11 ± 0.00 a | 0.05 ± 0.00 b | 0.03 ± 0.00 cd | 0.02 ± 0.00 e | 0.03 ± 0.00 d | 0.01 ± 0.00 fgh | 0.01 ± 0.00 i | 0.01 ± 0.00 hi | 0.01 ± 0.00 gh | 0.03 ± 0.00 c | 0.01 ± 0.00 hi | 0.03 ± 0.00 d |
| His | 0.02 ± 0.00 bc | 0.01 ± 0.00 d | 0.06 ± 0.01 a | 0.01 ± 0.00 cd | 0.01 ± 0.00 cd | 0.01 ± 0.00 cd | 0.01 ± 0.00 d | 0.01 ± 0.00 d | 0.02 ± 0.00 b | 0.01 ± 0.00 d | 0.01 ± 0.00 d | 0.01 ± 0.00 d | ND | 0.01 ± 0.00 d |
| Ile | 0.01 ± 0.00 de | 0.01 ± 0.00 efg | 0.03 ± 0.00 a | 0.01 ± 0.00 fg | 0.02 ± 0.00 cd | 0.01 ± 0.00 fg | 0.01 ± 0.00 fg | 0.01 ± 0.00 fg | 0.01 ± 0.00 def | 0.01 ± 0.00 gh | 0.02 ± 0.00 bc | 0.01 ± 0.00 gh | 0.01 ± 0.00 h | 0.02 ± 0.00 b |
| Arg | 0.01 ± 0.00 b | 0.02 ± 0.00 b | 1.73 ± 0.06 a | 0.03 ± 0.01 b | 0.01 ± 0.00 b | 0.01 ± 0.00 b | 0.01 ± 0.00 b | 0.02 ± 0.00 b | 0.03 ± 0.00 b | ND | 0.01 ± 0.00 b | 0.01 ± 0.00 b | ND | 0.02 ± 0.00 b |
| Asn | 0.01 ± 0.01 b | 0.01 ± 0.00 b | 0.16 ± 0.02 a | 0.02 ± 0.00 b | 0.01 ± 0.01 b | 0.01 ± 0.01 b | 0.01 ± 0.01 b | 0.01 ± 0.00 b | 0.03 ± 0.01 b | 0.01 ± 0.00 b | 0.01 ± 0.00 b | 0.01 ± 0.01 b | 0.01 ± 0.00 b | 0.01 ± 0.00 b |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Li, M.; Zheng, G.; Wang, T.; Lin, J.; Wang, S.; Wang, X.; Chao, Q.; Cao, S.; Yang, Z.; et al. Metabolite Profiling of 14 Wuyi Rock Tea Cultivars Using UPLC-QTOF MS and UPLC-QqQ MS Combined with Chemometrics. Molecules 2018, 23, 104. https://doi.org/10.3390/molecules23020104
Chen S, Li M, Zheng G, Wang T, Lin J, Wang S, Wang X, Chao Q, Cao S, Yang Z, et al. Metabolite Profiling of 14 Wuyi Rock Tea Cultivars Using UPLC-QTOF MS and UPLC-QqQ MS Combined with Chemometrics. Molecules. 2018; 23(2):104. https://doi.org/10.3390/molecules23020104
Chicago/Turabian StyleChen, Si, Meihong Li, Gongyu Zheng, Tingting Wang, Jun Lin, Shanshan Wang, Xiaxia Wang, Qianlin Chao, Shixian Cao, Zhenbiao Yang, and et al. 2018. "Metabolite Profiling of 14 Wuyi Rock Tea Cultivars Using UPLC-QTOF MS and UPLC-QqQ MS Combined with Chemometrics" Molecules 23, no. 2: 104. https://doi.org/10.3390/molecules23020104
APA StyleChen, S., Li, M., Zheng, G., Wang, T., Lin, J., Wang, S., Wang, X., Chao, Q., Cao, S., Yang, Z., & Yu, X. (2018). Metabolite Profiling of 14 Wuyi Rock Tea Cultivars Using UPLC-QTOF MS and UPLC-QqQ MS Combined with Chemometrics. Molecules, 23(2), 104. https://doi.org/10.3390/molecules23020104