Carbon Consumption Patterns of Microbial Communities Associated with Peltigera Lichens from a Chilean Temperate Forest
Abstract
:1. Introduction
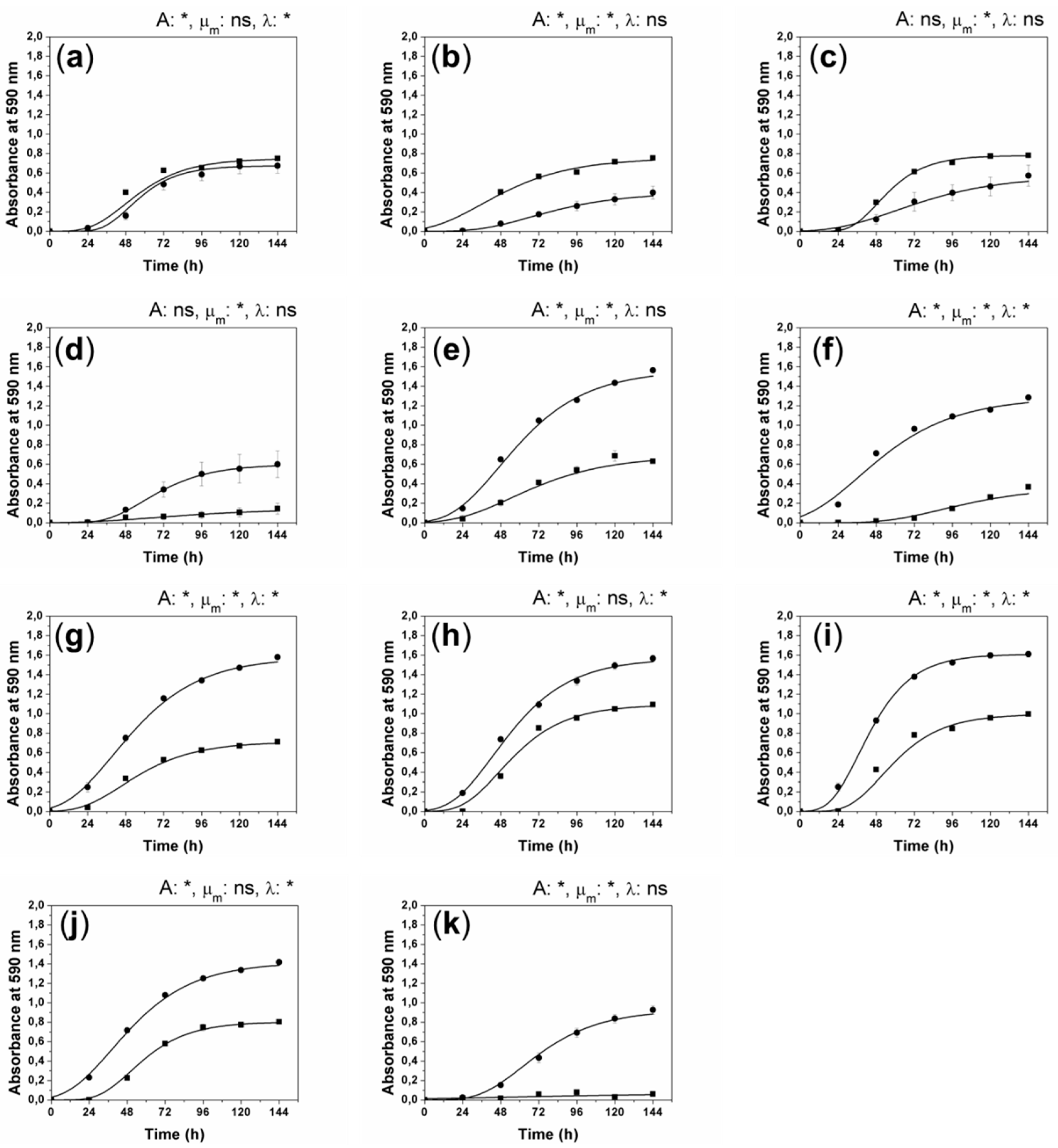
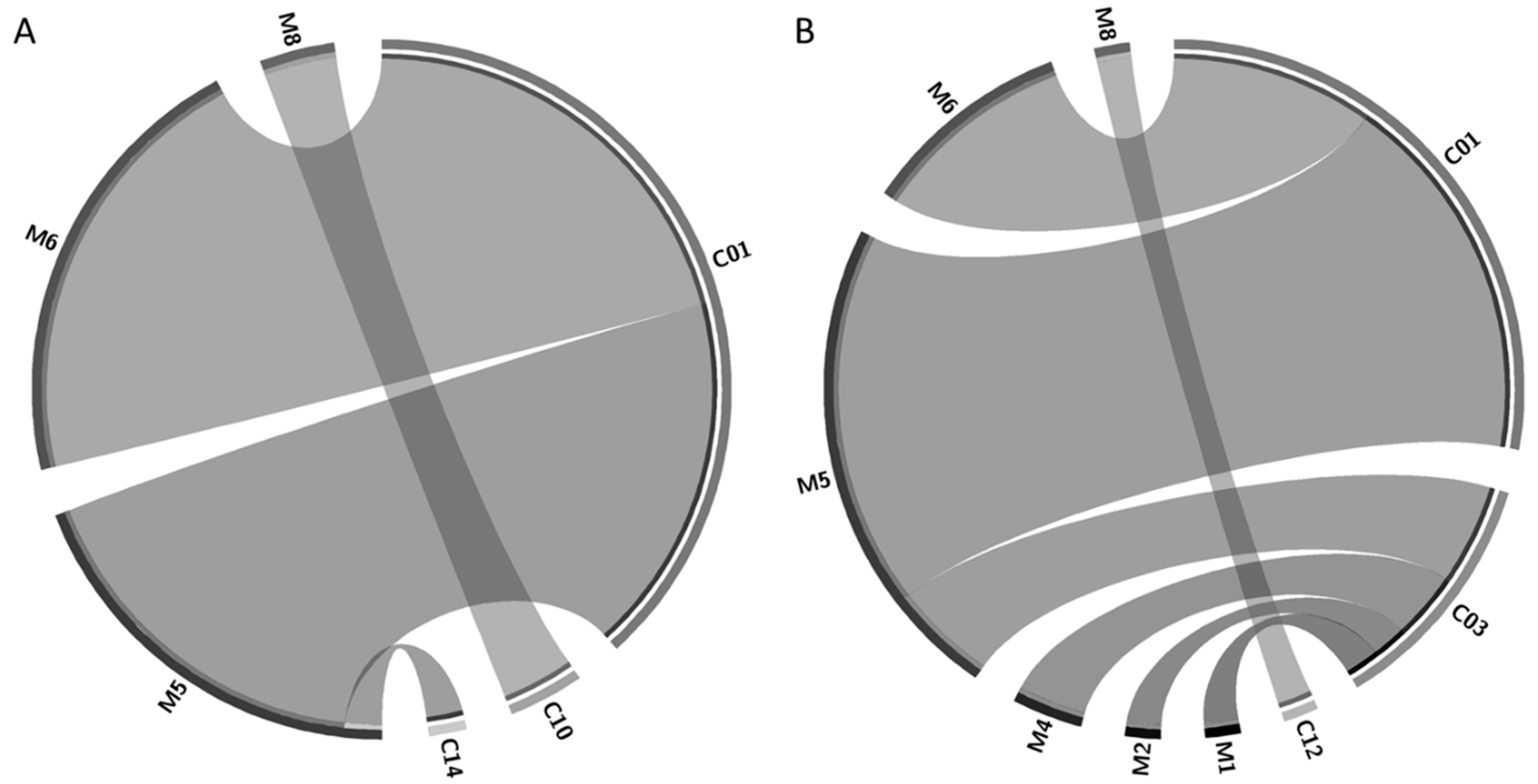
2. Results and Discussion
3. Materials and Methods
3.1. Study Sites and Sampling
3.2. Carbon-Source Utilization Pattern
3.3. Data Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Thalli | Substrates | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 h (S2M4C03-L) 1 | 72 h (S1M5C14-L) | 48 h (S2M5C03-S) | 72 h (S2M1C03-S) | ||||||||||
| Well | Carbon-Source | RSD 2 | Mean | CI 3 | RSD | Mean | CI | RSD | Mean | CI | RSD | Mean | CI |
| 1 | Water (blank) | 0.76 | 0.577 | 0.011 | 6.31 | 0.408 | 0.064 | 13.79 | 0.505 | 0.173 | 8.59 | 0.198 | 0.042 |
| 2 | Pyruvic acid methyl ester | 2.72 | 1.054 | 0.071 | 2.31 | 1.147 | 0.066 | 1.20 | 1.523 | 0.045 | 3.26 | 1.402 | 0.114 |
| 3 | Tween 40 | 1.85 | 1.261 | 0.058 | 23.41 | 1.520 | 0.884 | 5.58 | 2.011 | 0.279 | 5.24 | 1.604 | 0.209 |
| 4 | Tween 80 | 4.73 | 0.963 | 0.113 | 11.93 | 1.983 | 0.588 | 2.53 | 1.639 | 0.103 | 5.27 | 1.161 | 0.152 |
| 5 | α-cyclodextrin | 0.49 | 0.627 | 0.008 | 6.53 | 0.399 | 0.065 | 3.81 | 0.512 | 0.048 | 7.27 | 0.436 | 0.079 |
| 6 | Glycogen | 2.85 | 0.614 | 0.044 | 2.30 | 0.437 | 0.025 | 11.54 | 1.294 | 0.371 | 3.42 | 1.039 | 0.088 |
| 7 | d-cellobiose | 0.83 | 0.625 | 0.013 | 7.88 | 0.414 | 0.081 | 8.08 | 0.962 | 0.193 | 4.69 | 0.530 | 0.062 |
| 8 | α-d-lactose | 2.20 | 0.618 | 0.034 | 3.08 | 0.406 | 0.031 | 2.33 | 1.597 | 0.093 | 6.08 | 1.770 | 0.267 |
| 9 | β-methyl-d-glucoside | 2.16 | 0.604 | 0.032 | 7.04 | 0.408 | 0.071 | 3.25 | 2.024 | 0.164 | 1.33 | 1.977 | 0.065 |
| 10 | d-xylose | 1.85 | 1.572 | 0.072 | 11.23 | 0.706 | 0.197 | 1.47 | 1.677 | 0.061 | 5.90 | 1.414 | 0.207 |
| 11 | i-erythritol | 3.58 | 0.622 | 0.055 | 9.91 | 0.460 | 0.113 | 6.27 | 0.774 | 0.121 | 10.32 | 0.473 | 0.121 |
| 12 | d-mannitol | 1.37 | 1.473 | 0.050 | 2.92 | 3.016 | 0.219 | 1.30 | 2.383 | 0.077 | 5.18 | 2.045 | 0.263 |
| 13 | N-acetyl-d-glucosamine | 1.18 | 0.634 | 0.019 | 6.41 | 1.140 | 0.182 | 6.51 | 2.345 | 0.379 | 5.43 | 2.332 | 0.314 |
| 14 | d-glucosaminic acid | 0.87 | 1.367 | 0.030 | 11.71 | 0.594 | 0.173 | 1.02 | 1.523 | 0.039 | 4.63 | 1.831 | 0.210 |
| 15 | Glucose-1-phosphate | 2.70 | 0.715 | 0.048 | 7.63 | 0.447 | 0.085 | 11.65 | 1.530 | 0.443 | 2.06 | 1.119 | 0.057 |
| 16 | d.l-α-glycerol phosphate | 1.01 | 0.897 | 0.023 | 5.73 | 0.635 | 0.090 | 10.21 | 1.140 | 0.289 | 2.76 | 0.494 | 0.034 |
| 17 | d-galactonic acid-gamma-lactone | 3.39 | 1.723 | 0.145 | 7.88 | 1.465 | 0.287 | 1.96 | 1.581 | 0.077 | 5.82 | 1.860 | 0.269 |
| 18 | d-galacturonic acid | 2.92 | 2.280 | 0.166 | 7.70 | 0.807 | 0.154 | 11.80 | 2.135 | 0.626 | 5.01 | 2.245 | 0.279 |
| 19 | 2-Hydroxy benzoic acid | 5.42 | 0.167 | 0.023 | 5.73 | 0.107 | 0.015 | 1.17 | 1.702 | 0.049 | 2.57 | 0.243 | 0.016 |
| 20 | 4-Hydroxy benzoic acid | 1.52 | 0.497 | 0.019 | 3.57 | 1.788 | 0.159 | 3.59 | 1.559 | 0.139 | 5.04 | 1.588 | 0.199 |
| 21 | γ-hydroxybutyric acid | 6.12 | 0.758 | 0.115 | 6.60 | 0.477 | 0.078 | 16.53 | 0.680 | 0.279 | 16.72 | 0.352 | 0.146 |
| 22 | Itaconic acid | 4.39 | 0.986 | 0.107 | 2.35 | 0.419 | 0.024 | 3.65 | 1.780 | 0.161 | 2.71 | 2.311 | 0.156 |
| 23 | α-ketobutyric acid | 6.81 | 0.250 | 0.042 | 9.45 | 0.132 | 0.031 | 1.97 | 0.456 | 0.022 | 9.67 | 0.216 | 0.052 |
| 24 | d-malic acid | 1.65 | 0.708 | 0.029 | 6.07 | 0.564 | 0.085 | 8.97 | 0.513 | 0.114 | 4.07 | 0.877 | 0.089 |
| 25 | l-arginine | 6.76 | 0.864 | 0.145 | 6.31 | 2.071 | 0.325 | 5.84 | 2.145 | 0.311 | 1.36 | 2.649 | 0.090 |
| 26 | l-asparagine | 1.44 | 1.804 | 0.065 | 1.62 | 3.016 | 0.121 | 2.32 | 2.307 | 0.133 | 2.33 | 3.011 | 0.174 |
| 27 | l-phenylalanine | 3.04 | 0.639 | 0.048 | 11.42 | 0.214 | 0.061 | 12.50 | 0.484 | 0.150 | 11.89 | 0.318 | 0.094 |
| 28 | l-serine | 8.25 | 1.028 | 0.211 | 1.29 | 2.730 | 0.088 | 3.55 | 2.553 | 0.225 | 14.38 | 2.384 | 0.851 |
| 29 | l-threonine | 7.02 | 0.783 | 0.137 | 14.97 | 0.463 | 0.172 | 15.38 | 0.596 | 0.228 | 6.31 | 0.266 | 0.042 |
| 30 | Glycyl-l-glutamic acid | 1.44 | 1.096 | 0.039 | 1.45 | 1.448 | 0.052 | 8.15 | 0.996 | 0.202 | 12.22 | 0.701 | 0.213 |
| 31 | Phenylethylamine | 9.75 | 0.578 | 0.140 | 18.82 | 0.279 | 0.130 | 7.06 | 0.583 | 0.102 | 4.34 | 1.623 | 0.175 |
| 32 | Putrescin | 2.43 | 0.656 | 0.040 | 3.23 | 1.655 | 0.133 | 4.16 | 1.496 | 0.154 | 7.53 | 1.175 | 0.220 |
References
- Nash, T. Lichen Biology, 2nd ed.; Nash, T.H., Ed.; Cambridge University Press: Cambridge, UK, 2008; ISBN 9780511790478. [Google Scholar]
- Ainsworth, G.C. Dictionary of the Fungi, 10th ed.; Kirk, P.M., Cannon, P.F., Minter, D.W., Stalpers, J.A., Eds.; CABI Bioscience: Cambridge, UK, 2008; ISBN 9780851998268. [Google Scholar]
- Ahmadjian, V. Lichens are more important than you think. Bioscience 1995, 45, 124. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Callaghan, T.V.; Alatalo, J.M.; Michelsen, A.; Graglia, E.; Hartley, A.E.; Hik, D.S.; Hobbie, S.E.; Press, M.C.; Robinson, C.H.; et al. Global change and arctic ecosystems: Is lichen decline a function of increases in vascular plant biomass? J. Ecol. 2001, 89, 984–994. [Google Scholar] [CrossRef]
- Galloway, D.J. Lichen biogeography. In Lichen Biology; Nash, T.H., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 315–335. [Google Scholar]
- Miadlikowska, J.; Richardson, D.; Magain, N.; Ball, B.; Anderson, F.; Cameron, R.; Lendemer, J.; Truong, C.; Lutzoni, F. Phylogenetic placement, species delimitation, and cyanobiont identity of endangered aquatic Peltigera species (lichen-forming Ascomycota, Lecanoromycetes). Am. J. Bot. 2014, 101, 1141–1156. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.A. Weathering, secondary mineral genesis, and soil formation caused by lichens and mosses growing on granitic gneiss in a boreal forest environment. Geoderma 2015, 251–252, 78–91. [Google Scholar] [CrossRef]
- Beckett, R.P.; Zavarzina, A.G.; Liers, C. Oxidoreductases and cellulases in lichens: Possible roles in lichen biology and soil organic matter turnover. Fungal Biol. 2013, 117, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M. Geomycology: Biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 2007, 111, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Magain, N.; Truong, C.; Goward, T.; Niu, D.; Goffinet, B.; Sérusiaux, E.; Vitikainen, O.; Lutzoni, F.; Miadlikowska, J. Species delimitation at a global scale reveals high species richness with complex biogeography and patterns of symbiont association in Peltigera section Peltigera (lichenized Ascomycota: Lecanoromycetes). Taxon 2018. [Google Scholar] [CrossRef]
- Zúñiga, C.; Leiva, D.; Carú, M.; Orlando, J. Substrates of Peltigera lichens as a potential source of cyanobionts. Microb. Ecol. 2017, 74, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Magain, N.; Miadlikowska, J.; Coyle, J.R.; Truong, C.; Lutzoni, F. Bioclimatic factors at an intrabiome scale are more limiting than cyanobiont availability for the lichen-forming genus Peltigera. Am. J. Bot. 2018, 105, 1198–1211. [Google Scholar] [CrossRef] [PubMed]
- Jüriado, I.; Kaasalainen, U.; Rikkinen, J. Specialist taxa restricted to threatened habitats contribute significantly to the regional diversity of Peltigera (Lecanoromycetes, Ascomycota) in Estonia. Fungal Ecol. 2017, 30, 76–87. [Google Scholar] [CrossRef]
- Magain, N.; Miadlikowska, J.; Goffinet, B.; Sérusiaux, E.; Lutzoni, F. Macroevolution of specificity in cyanolichens of the genus Peltigera Section Polydactylon (Lecanoromycetes, Ascomycota). Syst. Biol. 2016, 66, syw065. [Google Scholar] [CrossRef] [PubMed]
- Chagnon, P.L.; Magain, N.; Miadlikowska, J.; Lutzoni, F. Strong specificity and network modularity at a very fine phylogenetic scale in the lichen genus Peltigera. Oecologia 2018, 187, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Vitikainen, O. Taxonomic Revision of Peltigera (Lichenized Ascomycotina) in Europe; Acta botanica Fennica; Finnish Zoological and Botanical Publishing Board: Helsinki, Finland, 1994; ISBN 9789519469461. [Google Scholar]
- Miadlikowska, J.; Lutzoni, F. Phylogenetic revision of the genus Peltigera (lichen-forming Ascomycota) based on morphological, chemical, and large subunit nuclear ribosomal DNA data. Int. J. Plant Sci. 2000, 161, 925–958. [Google Scholar] [CrossRef]
- Goffinet, B.; Hastings, R.I. Two new sorediate taxa of Peltigera. Lichenologist 1995, 27, 43–58. [Google Scholar] [CrossRef]
- Goffinet, B.; Miadlikowska, J.; Goward, T. Phylogenetic inferences based on nrDNA sequences support five morphospecies within the Peltigera didactyla complex (lichenized Ascomycota). Bryologist 2003, 106, 349–364. [Google Scholar] [CrossRef]
- Miadlikowska, J.; Lutzoni, F.; Goward, T.; Zoller, S.; Posada, D. New approach to an old problem: Incorporating signal from gap-rich regions of ITS and rDNA large subunit into phylogenetic analyses to resolve the Peltigera canina species complex. Mycologia 2003, 95, 1181–1203. [Google Scholar] [CrossRef] [PubMed]
- MartÍnez, I.; Burgaz, A.R.; Vitikainen, O.; Escudero, A. Distribution patterns in the genus Peltigera Willd. Lichenologist 2003, 35, 301–323. [Google Scholar] [CrossRef]
- Quilhot, W.; Cuellar, M.; Díaz, R.; Riquelme, F.; Rubio, C. Lichens of Aisen, Southern Chile. Gayana Bot. 2012, 69, 57–87. [Google Scholar] [CrossRef]
- Ramírez-Fernández, L.; Zúñiga, C.; Méndez, M.A.; Carú, M.; Orlando, J. Genetic diversity of terricolous Peltigera cyanolichen communities in different conservation states of native forest from southern Chile. Int. Microbiol. 2013, 16, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, C.; Leiva, D.; Ramírez-Fernández, L.; Carú, M.; Yahr, R.; Orlando, J. Phylogenetic diversity of Peltigera cyanolichens and their photobionts in Southern Chile and Antarctica. Microb. Environ. 2015, 30, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, I.A.; Cernava, T.; Berg, G.; Grube, M. Understanding microbial multi-species symbioses. Front. Microbiol. 2016, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Puglia, A.M.; Grube, M. Molecular analysis of lichen-associated bacterial communities. FEMS Microbiol. Ecol. 2006, 57, 484–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muggia, L.; Kopun, T.; Grube, M. Effects of growth media on the diversity of culturable fungi from lichens. Molecules 2017, 22, 824. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Cardinale, M.; de Castro, J., Jr.; Müller, H.; Berg, G.; de Castro, J.V.; Müller, H.; Berg, G. Species-specific structural and functional diversity of bacterial communities in lichen symbioses. ISME J. 2009, 3, 1105–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cernava, T.; Müller, H.; Aschenbrenner, I.A.; Grube, M.; Berg, G. Analyzing the antagonistic potential of the lichen microbiome against pathogens by bridging metagenomic with culture studies. Front. Microbiol. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sigurbjörnsdóttir, M.A.; Andrésson, Ó.S.; Vilhelmsson, O. Analysis of the Peltigera membranacea metagenome indicates that lichen-associated bacteria are involved in phosphate solubilization. Microbiology 2015, 161, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Sigurbjörnsdóttir, M.; Andrésson, Ó.S.; Vilhelmsson, O. Nutrient scavenging activity and antagonistic factors of non-photobiont lichen-associated bacteria: A review. World J. Microbiol. Biotechnol. 2016, 32, 68. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Cernava, T.; Soh, J.; Fuchs, S.; Aschenbrenner, I.; Lassek, C.; Wegner, U.; Becher, D.; Riedel, K.; Sensen, C.W.; et al. Exploring functional contexts of symbiotic sustain within lichen-associated bacteria by comparative omics. ISME J. 2015, 9, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Garland, J.L. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 1997, 24, 289–300. [Google Scholar] [CrossRef]
- Almasia, R.; Carú, M.; Handford, M.; Orlando, J. Environmental conditions shape soil bacterial community structure in a fragmented landscape. Soil Biol. Biochem. 2016, 103, 39–45. [Google Scholar] [CrossRef]
- Feigl, V.; Ujaczki, É.; Vaszita, E.; Molnár, M. Influence of red mud on soil microbial communities: Application and comprehensive evaluation of the Biolog EcoPlate approach as a tool in soil microbiological studies. Sci. Total Environ. 2017, 595, 903–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlando, J.; Chávez, M.; Bravo, L.; Guevara, R.; Carú, M. Effect of Colletia hystrix (Clos), a pioneer actinorhizal plant from the Chilean matorral, on the genetic and potential metabolic diversity of the soil bacterial community. Soil Biol. Biochem. 2007, 39, 2769–2776. [Google Scholar] [CrossRef]
- Green, J.L.; Bohannan, B.J.M.; Whitaker, R.J. Microbial biogeography: From taxonomy to traits. Science 2008, 320, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lennon, J.T. The generation and maintenance of diversity in microbial communities. Am. J. Bot. 2011, 98, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Grube, M.; Castro, J.V., Jr.; Müller, H.; Berg, G. Bacterial taxa associated with the lung lichen Lobaria pulmonaria are differentially shaped by geography and habitat. FEMS Microbiol. Lett. 2012, 329, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Hodkinson, B.P.; Gottel, N.R.; Schadt, C.W.; Lutzoni, F.F. Photoautotrophic symbiont and geography are major factors affecting highly structured and diverse bacterial communities in the lichen microbiome. Environ. Microbiol. 2012, 14, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Fernández, L.; Zúñiga, C.; Carú, M.; Orlando, J. Environmental context shapes the bacterial community structure associated to Peltigera cyanolichens growing in Tierra del Fuego, Chile. World J. Microbiol. Biotechnol. 2014, 30, 1141–1144. [Google Scholar] [CrossRef] [PubMed]
- Leiva, D.; Clavero-León, C.; Carú, M.; Orlando, J. Intrinsic factors of Peltigera lichens influence the structure of the associated soil bacterial microbiota. FEMS Microbiol. Ecol. 2016, 92, fiw178. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, H.E.; Miadlikowska, J.; Lutzoni, F. Assessing reproductive isolation in highly diverse communities of the lichen-forming fungal genus Peltigera. Evolution 2009, 63, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Miadlikowska, J.; Kauff, F.; Hofstetter, V.; Fraker, E.; Grube, M.; Hafellner, J.; Reeb, V.; Hodkinson, B.P.; Kukwa, M.; Lücking, R.; et al. New insights into classification and evolution of the Lecanoromycetes (Pezizomycotina, Ascomycota) from phylogenetic analyses of three ribosomal RNA- and two protein-coding genes. Mycologia 2006, 98, 1088–1103. [Google Scholar] [CrossRef] [PubMed]
- Miadlikowska, J.; Kauff, F.; Högnabba, F.; Oliver, J.C.; Molnár, K.; Fraker, E.; Gaya, E.; Hafellner, J.; Hofstetter, V.; Gueidan, C.; et al. A multigene phylogenetic synthesis for the class Lecanoromycetes (Ascomycota): 1307 fungi representing 1139 infrageneric taxa, 317 genera and 66 families. Mol. Phylogenet. Evol. 2014, 79, 132–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastida, F.; Jehmlich, N.; Ondoño, S.; von Bergen, M.; García, C.; Moreno, J.L. Characterization of the microbial community in biological soil crusts dominated by Fulgensia desertorum (Tomin) Poelt and Squamarina cartilaginea (With.) P. James and in the underlying soil. Soil Biol. Biochem. 2014, 76, 70–79. [Google Scholar] [CrossRef]
- De Armas-Ricard, M.; Orlando, J.; Bustamante, R.; Carú, M. Microbial communities of bulk and Eschscholzia californica rhizosphere soils at two altitudes in Central Chile. J. Soil Sci. Plant Nutr. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Till-Bottraud, I.; Fajardo, A.; Rioux, D. Multi-stemmed trees of Nothofagus pumilio second-growth forest in Patagonia are formed by highly related individuals. Ann. Bot. 2012, 110, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Docherty, K.M.; Borton, H.M.; Espinosa, N.; Gebhardt, M.; Gil-Loaiza, J.; Gutknecht, J.L.M.; Maes, P.W.; Mott, B.M.; Parnell, J.J.; Purdy, G.; et al. Key edaphic properties largely explain temporal and geographic variation in soil microbial communities across four biomes. PLoS ONE 2015, 10, e0135352. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Gallardo, A.; Covelo, F.; Prado-Comesaña, A.; Ochoa, V.; Maestre, F.T. Differences in thallus chemistry are related to species-specific effects of biocrust-forming lichens on soil nutrients and microbial communities. Funct. Ecol. 2015, 29, 1087–1098. [Google Scholar] [CrossRef] [Green Version]
- Song, S.H.; Vieille, C. Recent advances in the biological production of mannitol. Appl. Microbiol. Biotechnol. 2009, 84, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Solomon, P.S.; Waters, O.D.C.; Oliver, R.P. Decoding the mannitol enigma in filamentous fungi. Trends Microbiol. 2007, 15, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Kidron, G.J.; Pen-Mouratov, S.; Wasserstrom, H.; Barness, G.; Steinberger, Y. Do development stages of biological soil crusts determine activity and functional diversity in a sand-dune ecosystem? Soil Biol. Biochem. 2012, 51, 66–72. [Google Scholar] [CrossRef]
- Hamer, U.; Marschner, B. Priming effects in soils after combined and repeated substrate additions. Geoderma 2005, 128, 38–51. [Google Scholar] [CrossRef]
- Orwin, K.H.; Wardle, D.A.; Greenfield, L.G. Ecological consequences of carbon substrate identity and diversity in a laboratory study. Ecology 2006, 87, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Vranova, V.; Rejsek, K.; Skene, K.R.; Formanek, P. Non-protein amino acids: Plant, soil and ecosystem interactions. Plant Soil 2011, 342, 31–48. [Google Scholar] [CrossRef]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Heberer, T. Occurrence, fate, and removal of pharmaceutical residues in the aquatic environment: A review of recent research data. Toxicol. Lett. 2002, 131, 5–17. [Google Scholar] [CrossRef]
- Guinea, E.; Arias, C.; Cabot, P.L.; Garrido, J.A.; Rodríguez, R.M.; Centellas, F.; Brillas, E. Mineralization of salicylic acid in acidic aqueous medium by electrochemical advanced oxidation processes using platinum and boron-doped diamond as anode and cathodically generated hydrogen peroxide. Water Res. 2008, 42, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Anorve, L.I.; Calcagno, M.L.; Plumbridge, J. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J. Bacteriol. 2005, 187, 2974–2982. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Rodionov, D.A.; Li, X.; Laikova, O.N.; Gelfand, M.S.; Zagnitko, O.P.; Romine, M.F.; Obraztsova, A.Y.; Nealson, K.H.; Osterman, A.L. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 2006, 281, 29872–29885. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Hilditch, S. D-galacturonic acid catabolism in microorganisms and its biotechnological relevance. Appl. Microbiol. Biotechnol. 2009, 82, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Huang, X.; Zhong, C.; Li, J.; Lu, X. Identification of an itaconic acid degrading pathway in itaconic acid producing Aspergillus terreus. Appl. Microbiol. Biotechnol. 2016, 100, 7541–7548. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Byun, B.Y.; Mah, J.-H. Biogenic amine formation and bacterial contribution in Natto products. Food Chem. 2012, 135, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Güven, K.C.; Percot, A.; Sezik, E. Alkaloids in marine algae. Mar. Drugs 2010, 8, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grohmann, K.; Baldwin, E.A.; Buslig, B.S.; Ingram, L.O. Fermentation of galacturonic acid and other sugars in orange peel hydrolysates by the ethanologenic strain of Escherichia coli. Biotechnol. Lett. 1994, 16, 281–286. [Google Scholar] [CrossRef]
- Grohmann, K.; Manthey, J.A.; Cameron, R.G.; Buslig, B.S. Fermentation of galacturonic acid and pectin-rich materials to ethanol by genetically modified strains of Erwinia. Biotechnol. Lett. 1998, 20, 195–200. [Google Scholar] [CrossRef]
- Steiger, M.G.; Blumhoff, M.L.; Mattanovich, D.; Sauer, M. Biochemistry of microbial itaconic acid production. Front. Microbiol. 2013, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, I.A.; Cernava, T.; Erlacher, A.; Berg, G.; Grube, M. Differential sharing and distinct co-occurrence networks among spatially close bacterial microbiota of bark, mosses and lichens. Mol. Ecol. 2017, 26, 2826–2838. [Google Scholar] [CrossRef] [PubMed]
- Boustie, J.; Grube, M. Lichens-a promising source of bioactive secondary metabolites. Plant Genet. Resour. Charact. Util. 2005, 3, 273–287. [Google Scholar] [CrossRef]
- Salgado, F.; Albornoz, L.; Cortéz, C.; Stashenko, E.; Urrea-Vallejo, K.; Nagles, E.; Galicia-Virviescas, C.; Cornejo, A.; Ardiles, A.; Simirgiotis, M.; et al. Secondary metabolite profiling of species of the genus Usnea by UHPLC-ESI-OT-MS-MS. Molecules 2017, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- White, P.A.S.; Oliveira, R.C.M.; Oliveira, A.P.; Serafini, M.R.; Araújo, A.A.S.; Gelain, D.P.; Moreira, J.C.F.; Almeida, J.R.G.S.; Quintans, J.S.S.; Quintans-Junior, L.J.; et al. V Antioxidant activity and mechanisms of action of natural compounds isolated from lichens: A systematic review. Molecules 2014, 19, 14496–14527. [Google Scholar] [CrossRef] [PubMed]
- Legouin, B.; Lohézic-Le Dévéhat, F.; Ferron, S.; Rouaud, I.; Le Pogam, P.; Cornevin, L.; Bertrand, M.; Boustie, J. Specialized metabolites of the lichen Vulpicida pinastri act as photoprotective agents. Molecules 2017, 22, 1162. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.-S.; Kim, T.; Kim, J.; Hong, J.-M.; Nguyen, T.; Han, S.; Youn, U.; Yim, J.; Kim, I.-C. Anticancer activity of ramalin, a secondary metabolite from the antarctic lichen Ramalina terebrata, against colorectal cancer cells. Molecules 2017, 22, 1361. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.-M.; Suh, S.-S.; Kim, T.; Kim, J.; Han, S.; Youn, U.; Yim, J.; Kim, I.-C. Anti-cancer activity of lobaric acid and lobarstin extracted from the antarctic lichen Stereocaulon alpnum. Molecules 2018, 23, 658. [Google Scholar] [CrossRef] [PubMed]
- Parrot, D.; Legrave, N.; Delmail, D.; Grube, M.; Suzuki, M.; Tomasi, S. Review—Lichen-associated bacteria as a hot spot of chemodiversity: Focus on uncialamycin, a promising compound for future medicinal applications. Planta Med. 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nicolardi, V.; Cai, G.; Parrotta, L.; Puglia, M.; Bianchi, L.; Bini, L.; Gaggi, C. The adaptive response of lichens to mercury exposure involves changes in the photosynthetic machinery. Environ. Pollut. 2012, 160, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Parrot, D.; Legrave, N.; Intertaglia, L.; Rouaud, I.; Legembre, P.; Grube, M.; Suzuki, M.T.; Tomasi, S. Cyaneodimycin, a bioactive compound isolated from the culture of Streptomyces cyaneofuscatus associated with Lichina confinis. Eur. J. Org. Chem. 2016, 2016, 3977–3982. [Google Scholar] [CrossRef]
- Parrot, D.; Intertaglia, L.; Jehan, P.; Grube, M.; Suzuki, M.T.; Tomasi, S. Chemical analysis of the Alphaproteobacterium strain MOLA1416 associated with the marine lichen Lichina pygmaea. Phytochemistry 2018, 145, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Noël, A.; Ferron, S.; Rouaud, I.; Gouault, N.; Hurvois, J.-P.; Tomasi, S. Isolation and structure identification of novel brominated diketopiperazines from Nocardia ignorata—A lichen-associated Actinobacterium. Molecules 2017, 22, 371. [Google Scholar] [CrossRef] [PubMed]
- Wilmotte, A.; Van der Auwera, G.; De Wachter, R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef]
- Insam, H.; Goberna, M. Use of Biolog® for the Community Level Physiological Profiling (CLPP) of environmental samples. In Molecular Microbial Ecology Manual; Kowalchuk, G.A., de Bruijn, F.J., Head, I.M., Akkermans, A.D., van Elsas, J.D., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 853–860. ISBN 1-4020-2176-3. [Google Scholar]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, Ø.; Harper, D.; Ryan, P. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]

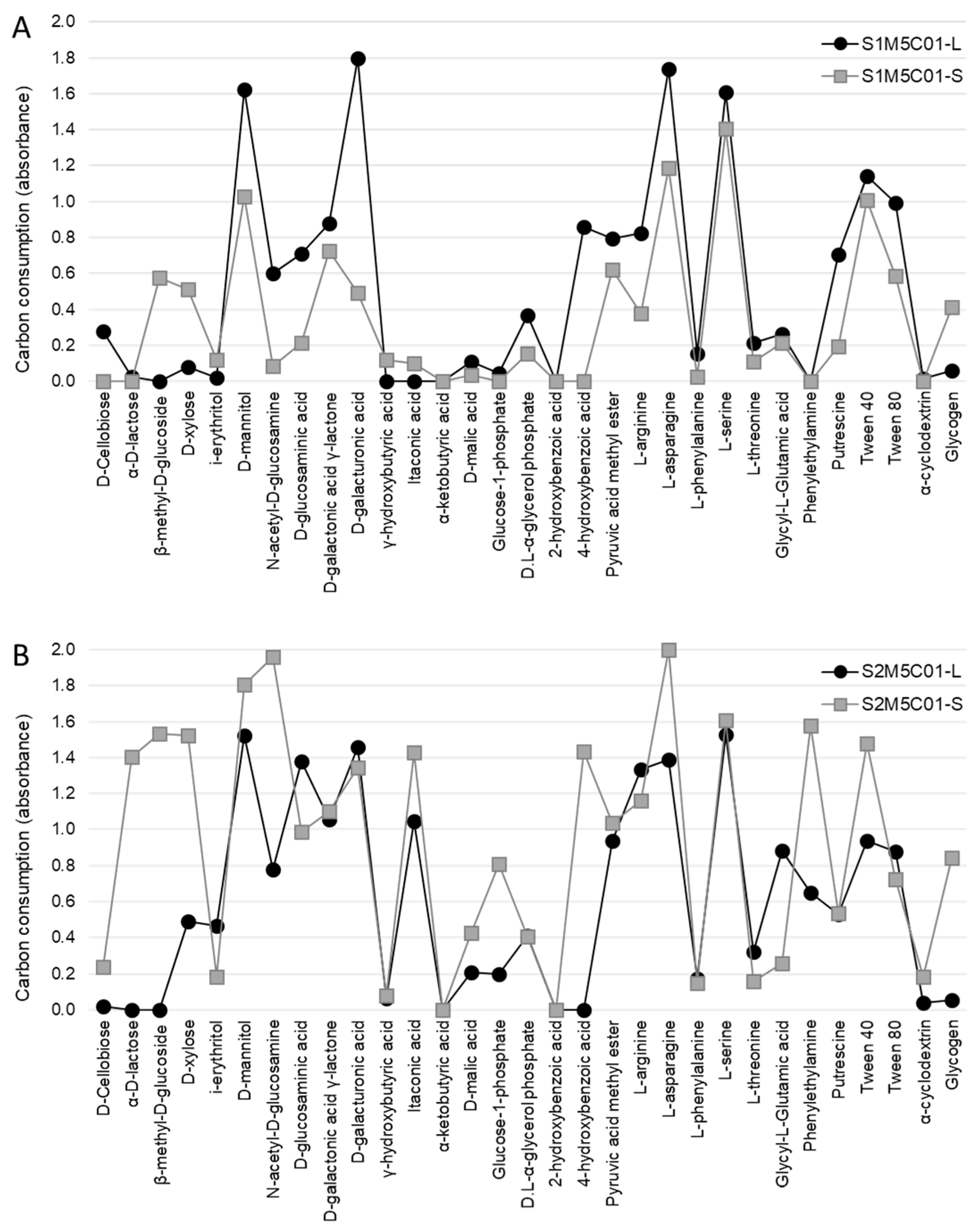

| S1M5C01-L | S1M5C01-S | S1M6C01-L | S1M6C01-S | S2M5C01-L | S2M5C01-S | S2M6C01-L | S2M6C01-S | |
|---|---|---|---|---|---|---|---|---|
| Carbohydrates | ||||||||
| d-Cellobiose | 0.28 (0.43) A | 0.00 (0.00) A | 0.07 (0.08) A | 0.02 (0.05) A | 0.02 (0.04) A | 0.24 (0.18) A | 0.11 (0.09) A | 0.10 (0.11) A |
| α-d-lactose | 0.02 (0.06) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 1.40 (0.65) B | 0.00 (0.00) A | 0.06 (0.09) A |
| β-methyl-d-glucoside | 0.00 (0.00) A | 0.53 (0.87) AB | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 1.54 (0.22) B | 0.08 (0.09) A | 1.07 (0.61) AB |
| d-xylose | 0.08 (0.09) AB | 0.45 (0.79) AB | 0.17 (0.09) AB | 0.00 (0.00) A | 0.49 (0.35) AB | 1.52 (0.49) C | 0.35 (0.23) AB | 0.87 (0.41) BC |
| i-erythritol | 0.02 (0.05) A | 0.12 (0.19) A | 0.02 (0.06) A | 0.05 (0.07) A | 0.47 (0.43) A | 0.18 (0.22) A | 0.08 (0.06) A | 0.58 (0.36) A |
| d-mannitol | 1.62 (0.41) A | 1.00 (0.73) A | 1.63 (0.62) A | 0.77 (0.75) A | 1.52 (0.53) A | 1.80 (0.22) A | 1.03 (0.73) A | 2.00 (0.00) A |
| N-acetyl-d-glucosamine | 0.60 (0.40) AB | 0.09 (0.21) A | 0.53 (0.11) AB | 0.30 (0.16) AB | 0.78 (0.42) AB | 1.96 (0.06) C | 0.38 (0.11) AB | 1.30 (0.61) BC |
| Carboxylic acids | ||||||||
| d-glucosaminic acid | 0.71 (0.53) AB | 0.21 (0.39) A | 0.46 (0.45) AB | 0.84 (0.72) AB | 1.38 (0.49) B | 0.99 (0.35) AB | 0.72 (0.53) AB | 1.17 (0.47) BC |
| d-galactonic acid γ-lactone | 0.88 (0.04) B | 0.73 (0.55) AB | 0.63 (0.37) AB | 0.12 (0.17) A | 1.06 (0.20) BC | 1.10 (0.23) BC | 0.59 (0.24) AB | 1.53 (0.28) C |
| d-galacturonic acid | 1.80 (0.05) C | 0.49 (0.52) A | 0.61 (0.46) A | 0.63 (0.79) AB | 1.46 (0.59) AB | 1.35 (0.14) AB | 1.54 (0.51) AB | 1.60 (0.09) B |
| γ-hydroxybutyric acid | 0.00 (0.00) A | 0.12 (0.18) A | 0.04 (0.07) A | 0.06 (0.09) A | 0.12 (0.10) A | 0.10 (0.12) A | 0.04 (0.07) A | 0.07 (0.07) A |
| Itaconic acid | 0.00 (0.00) A | 0.10 (0.18) A | 0.10 (0.24) A | 0.32 (0.35) A | 1.05 (0.57) AB | 1.43 (0.43) B | 0.23 (0.44) A | 1.59 (0.36) B |
| α-ketobutyric acid | 0.00 (0.00) A | 0.00 (0.00) A | 0.05 (0.13) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A |
| d-malic acid | 0.00 (0.00) A | 0.03 (0.08) A | 0.11 (0.09) A | 0.00 (0.00) A | 0.21 (0.23) A | 0.43 (0.43) A | 0.06 (0.10) A | 0.39 (0.21) A |
| Phosphorylated chemicals | ||||||||
| Glucose-1-phosphate | 0.05 (0.08) A | 0.00 (0.00) A | 0.03 (0.07) A | 0.00 (0.00) A | 0.20 (0.12) AB | 0.81 (0.18) C | 0.10 (0.11) AB | 0.34 (0.17) B |
| d.l-α-glycerol phosphate | 0.37 (0.06) AB | 0.15 (0.20) A | 0.24 (0.06) AB | 0.15 (0.14) A | 0.41 (0.08) B | 0.41 (0.15) B | 0.29 (0.12) AB | 0.30 (0.05) AB |
| Aromatic compounds | ||||||||
| 2-hydroxybenzoic acid | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A |
| 4-hydroxybenzoic acid | 0.86 (0.77) BC | 0.00 (0.00) A | 0.54 (0.60) AB | 0.00 (0.00) A | 0.00 (0.00) A | 1.43 (0.31) C | 0.38 (0.42) AB | 1.12 (0.39) BC |
| Esters | ||||||||
| Pyruvic acid methyl ester | 0.79 (0.07) AB | 0.62 (0.61) AB | 0.47 (0.20) A | 0.40 (0.47) A | 0.94 (0.37) BC | 1.04 (0.20) BC | 0.49 (0.32) AB | 1.35 (0.21) C |
| Amino acids | ||||||||
| l-arginine | 0.82 (0.38) B | 0.38 (0.48) AB | 1.03 (0.79) B | 0.09 (0.17) A | 1.34 (0.73) B | 1.16 (0.57) B | 0.49 (0.38) AB | 1.44 (0.62) B |
| l-asparagine | 1.74 (0.29) B | 1.03 (0.81) AB | 1.40 (0.63) AB | 0.51 (0.73) A | 1.39 (0.68) AB | 2.00 (0.00) B | 1.29 (0.63) AB | 1.94 (0.07) B |
| l-phenylalanine | 0.16 (0.09) A | 0.03 (0.06) A | 0.07 (0.08) A | 0.00 (0.00) A | 0.17 (0.16) A | 0.15 (0.13) A | 0.11 (0.09) A | 0.10 (0.08) A |
| l-serine | 1.61 (0.13) A | 1.36 (0.63) A | 1.98 (0.04) A | 0.95 (0.90) A | 1.53 (0.47) A | 1.61 (0.27) A | 1.42 (0.58) A | 2.00 (0.00) A |
| l-threonine | 0.22 (0.07) A | 0.11 (0.13) A | 0.02 (0.04) A | 0.03 (0.08) A | 0.32 (0.26) A | 0.16 (0.14) A | 0.11 (0.14) A | 0.12 (0.09) A |
| Glycyl-l-Glutamic acid | 0.27 (0.08) A | 0.22 (0.36) A | 0.67 (0.27) AB | 0.24 (0.41) A | 0.88 (0.47) B | 0.26 (0.22) A | 0.25 (0.16) A | 0.45 (0.16) AB |
| Amines | ||||||||
| Phenylethylamine | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.00 (0.00) A | 0.65 (0.23) B | 1.58 (0.27) C | 0.00 (0.00) A | 1.79 (0.26) C |
| Putrescine | 0.70 (0.24) A | 0.19 (0.30) A | 1.01 (0.22) B | 0.38 (0.47) A | 0.53 (0.46) A | 0.54 (0.39) A | 0.73 (0.40) AB | 1.30 (0.09) B |
| Polymers | ||||||||
| Tween 40 | 1.14 (0.45) B | 1.01 (0.48) B | 0.96 (0.63) B | 0.35 (0.38) A | 0.94 (0.42) B | 1.48 (0.16) B | 0.58 (0.32) A | 1.17 (0.22) B |
| Tween 80 | 1.00 (0.35) A | 0.59 (0.62) A | 1.14 (0.62) A | 0.39 (0.43) A | 0.88 (0.31) A | 0.72 (0.22) A | 0.88 (0.52) A | 1.08 (0.33) A |
| α-cyclodextrin | 0.02 (0.04) A | 0.00 (0.00) A | 0.06 (0.07) A | 0.02 (0.05) A | 0.04 (0.10) A | 0.18 (0.20) A | 0.05 (0.08) A | 0.02 (0.04) A |
| Glycogen | 0.06 (0.09) A | 0.41 (0.43) AB | 0.12 (0.07) A | 0.04 (0.06) A | 0.05 (0.13) A | 0.84 (0.23) B | 0.12 (0.10) A | 1.00 (0.36) B |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almendras, K.; Leiva, D.; Carú, M.; Orlando, J. Carbon Consumption Patterns of Microbial Communities Associated with Peltigera Lichens from a Chilean Temperate Forest. Molecules 2018, 23, 2746. https://doi.org/10.3390/molecules23112746
Almendras K, Leiva D, Carú M, Orlando J. Carbon Consumption Patterns of Microbial Communities Associated with Peltigera Lichens from a Chilean Temperate Forest. Molecules. 2018; 23(11):2746. https://doi.org/10.3390/molecules23112746
Chicago/Turabian StyleAlmendras, Katerin, Diego Leiva, Margarita Carú, and Julieta Orlando. 2018. "Carbon Consumption Patterns of Microbial Communities Associated with Peltigera Lichens from a Chilean Temperate Forest" Molecules 23, no. 11: 2746. https://doi.org/10.3390/molecules23112746





