Reference Gene Selection for Quantitative Real-Time Reverse-Transcriptase PCR in Annual Ryegrass (Lolium multiflorum) Subjected to Various Abiotic Stresses
Abstract
:1. Introduction
2. Results
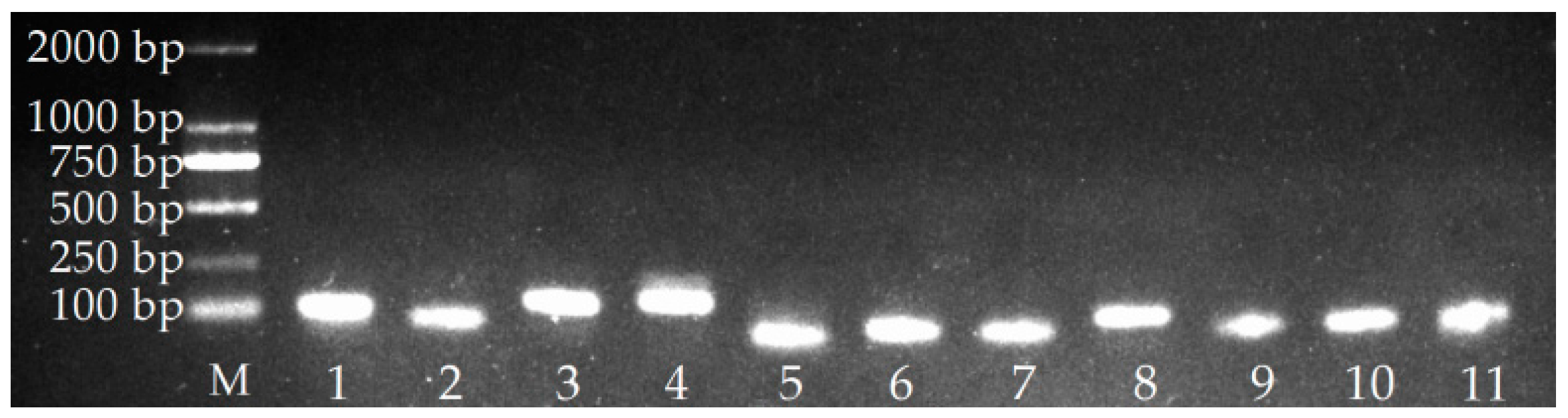
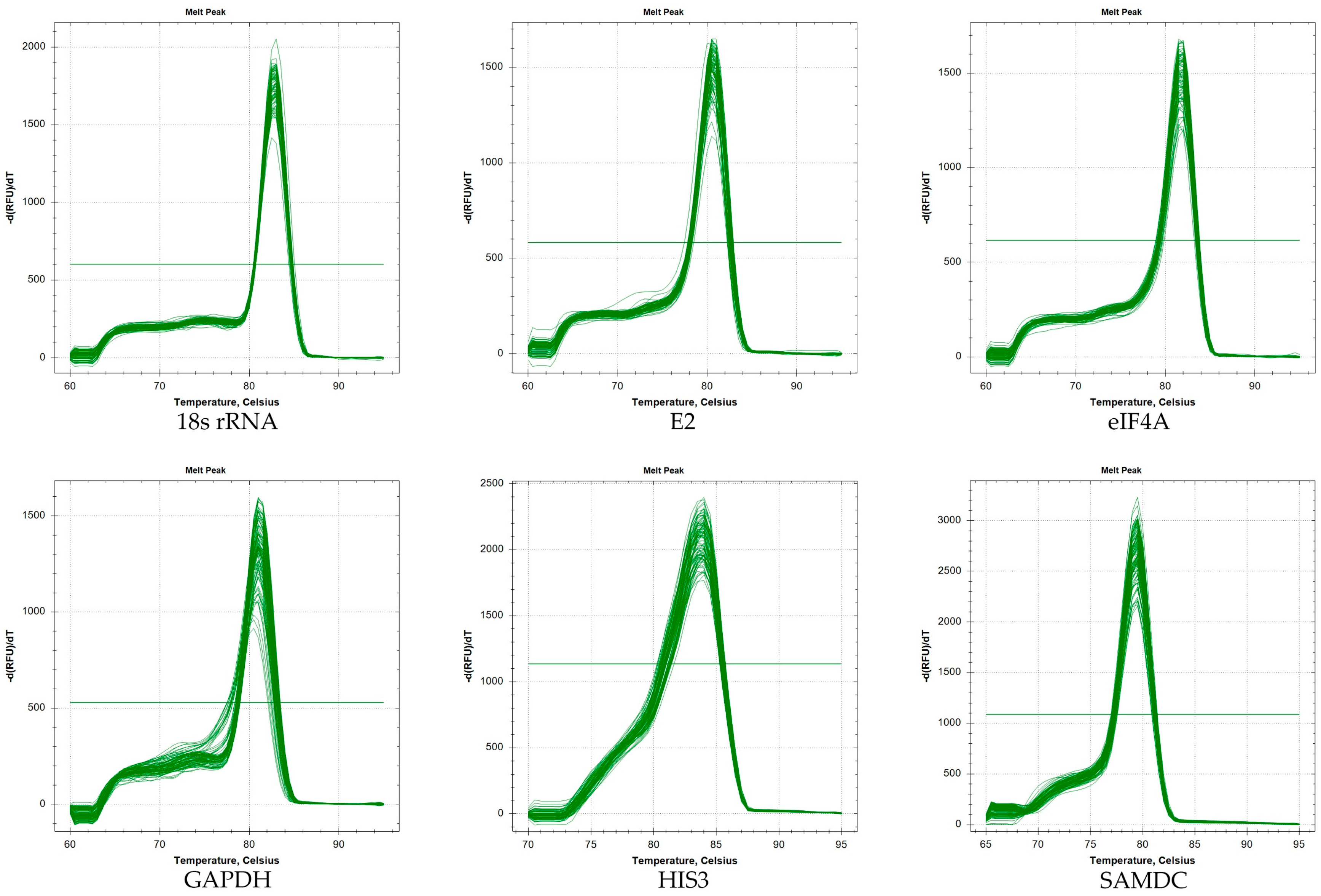
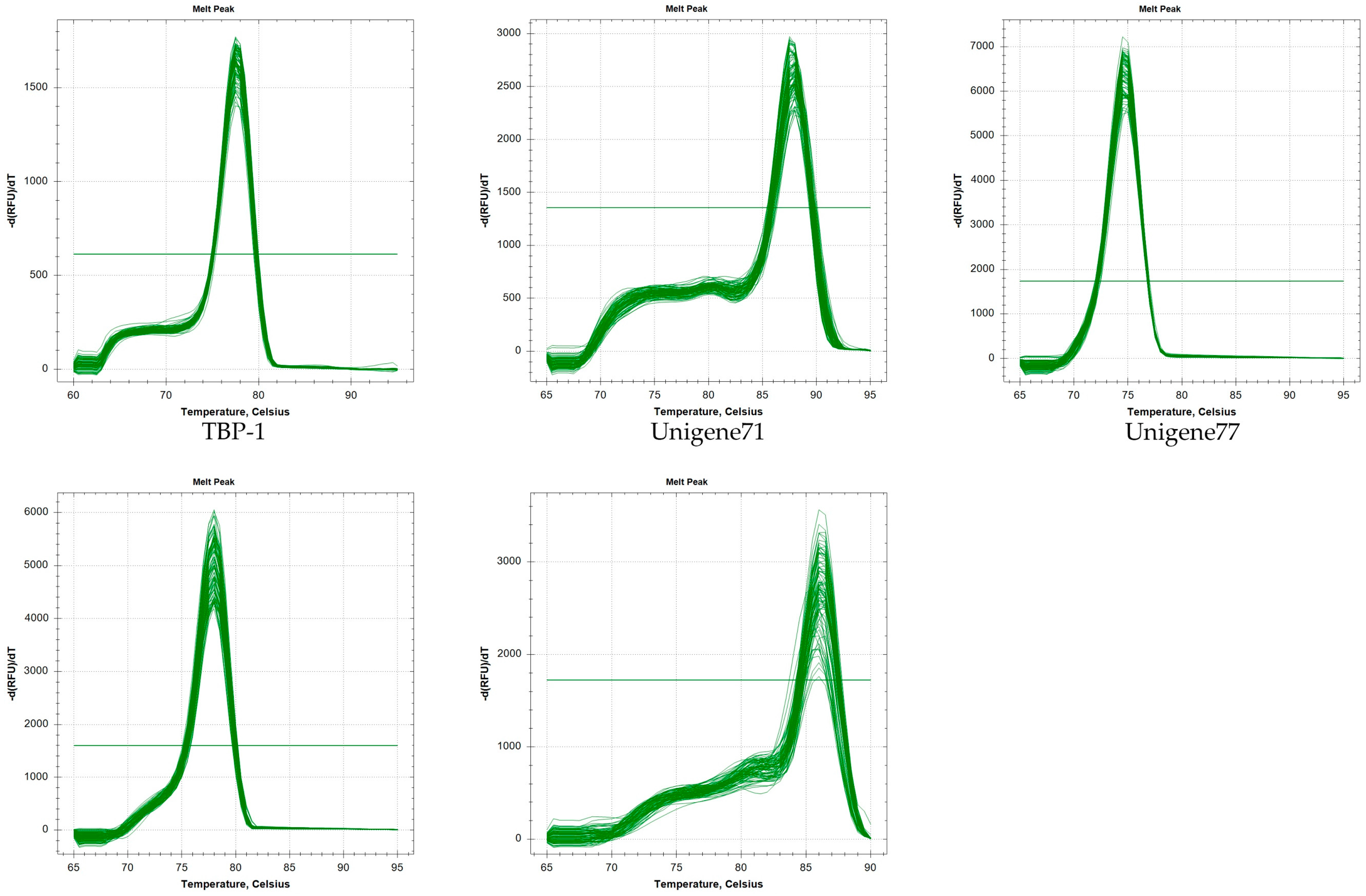
2.1. Verification of PCR Amplicons, Primer Specificity, and Gene-Specific PCR Amplification Efficiency
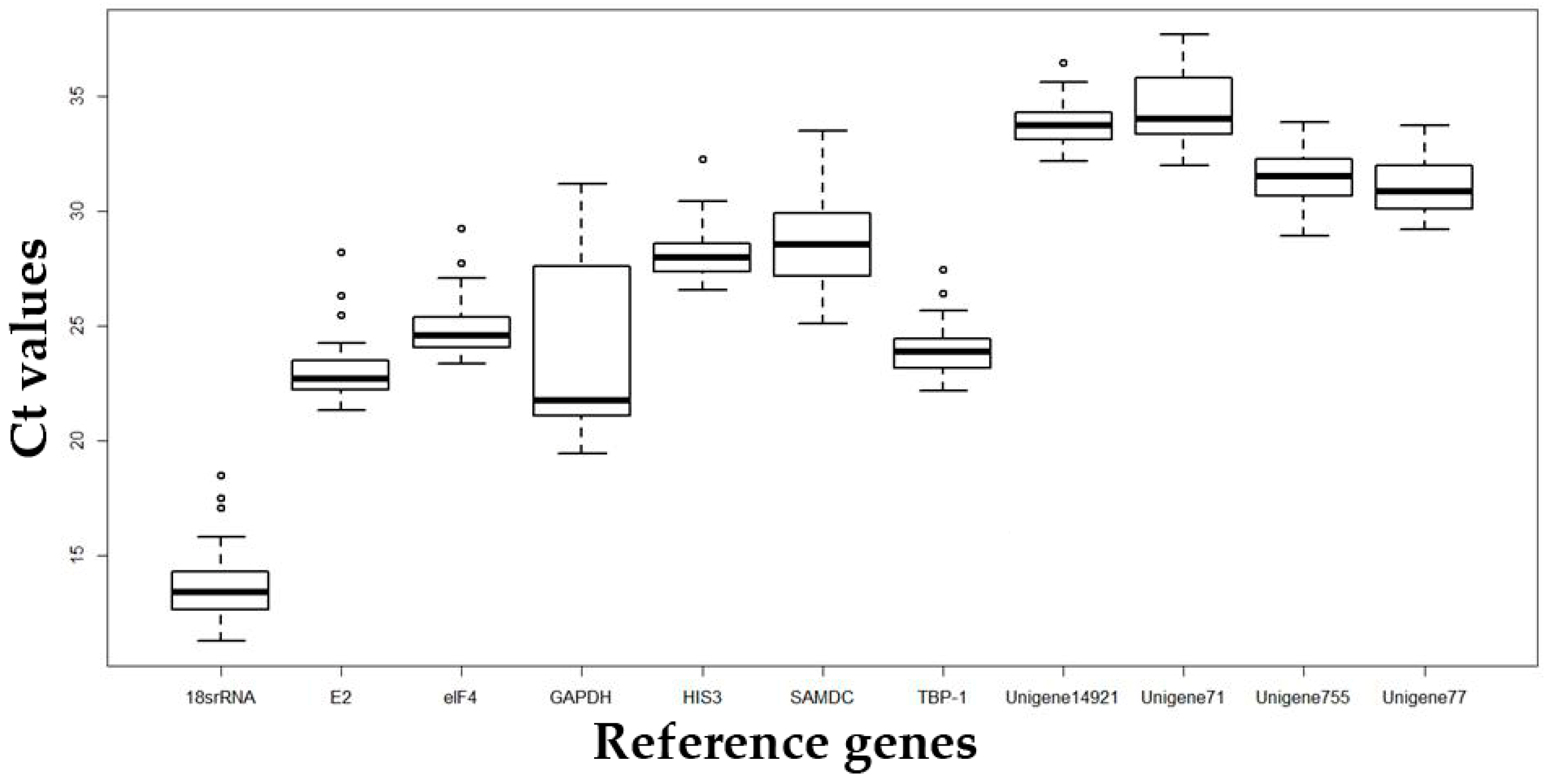
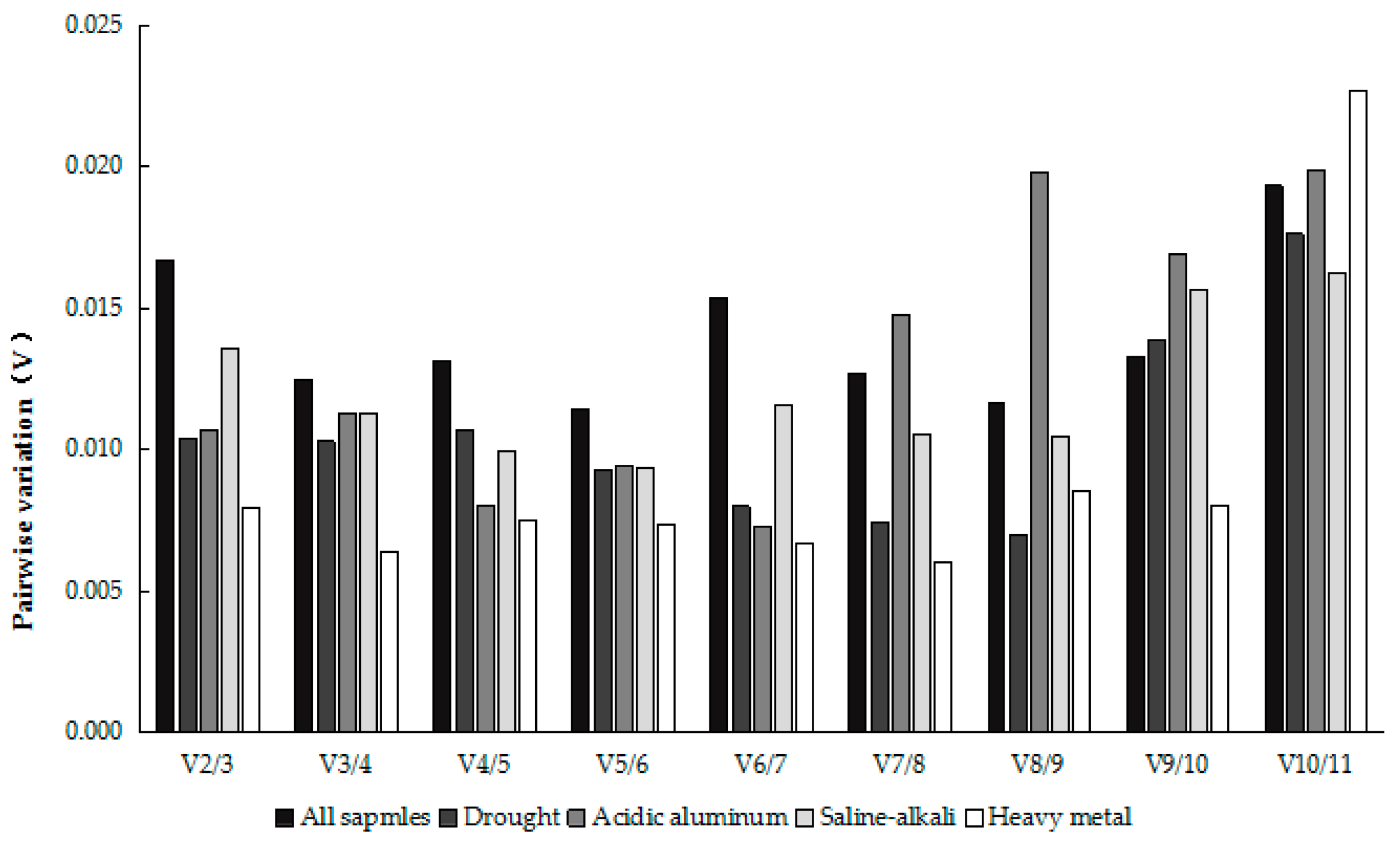
2.2. Analysis of the Expression Stability of Reference Genes under Different Types of Abiotic Stress
2.3. Stability Ranking of Candidate Reference Genes for Different Abiotic Stresses
2.3.1. GeNorm Analysis
2.3.2. BestKeeper Analysis
2.3.3. NormFinder Analysis
2.3.4. RefFinder Analysis
2.4. Validation of the Usefulness of the Reference Genes Identified from This Study
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Reagents and Instruments
4.3. RNA Extraction and First-Strand cDNA Synthesis
4.4. Selection of Reference Genes and PCR Primer Design
4.5. qRT-PCR Amplification Procedure
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Castanheira, N.; Dourado, A.C.; Alves, P.I.; Cortéspallero, A.M.; Delgadorodríguez, A.I.; Prazeres, Â.; Borges, N.; Sánchez, C.; Barreto Crespo, M.T.; Fareleira, P. Annual ryegrass-associated bacteria with potential for plant growth promotion. Microbiol. Res. 2014, 169, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, H.; Lin, C. Physiological, biochemical and growth responses of Italian ryegrass to butachlor exposure. Pestic. Biochem. Physiol. 2013, 106, 21–27. [Google Scholar] [CrossRef]
- Zeng, K. Study on Genetic Diversity of Germplasm Resources of Lolium L. J. Anhui Agric. Sci. 2007, 35, 3252–3254. [Google Scholar]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Relative quantification. In Real-Time PCR; Dorak, T., Ed.; Taylor and Francis Group: New York, NY, USA, 2006; pp. 63–82. [Google Scholar]
- Quackenbush, J. Microarray data normalization and transformation. Nat. Genet. 2002, 32, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Schöffl, F. Heat-stress-dependency and developmental modulation of gene expression: The potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR. J. Exp. Bot. 2003, 54, 2343–2349. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan, P.H.; Teng, K.; Yan, M.J.; He, C.Y.; Gan, L.; Yin, S.X.; Zhang, L.; Tan, P.H.; Teng, K. Screening of reference genes for real-time fluorescence quantitative PCR in Kentucky bluegrass. Acta Pratacult. Sin. 2017, 3, 75–81. [Google Scholar]
- Hong, S.Y.; Seo, P.J.; Yang, M.S.; Xiang, F.; Park, C.M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Qian, Y.; Han, L.; Sun, Z.; Fan, C.; Liu, J.; Ju, G. Selection of reliable reference genes for gene expression studies in Rhododendron micranthum Turcz. Sci. Hortic. 2012, 138, 128–133. [Google Scholar] [CrossRef]
- Sun, Y.L.; Zhang, D.H.; Zhao, L.; Xia, C.C.; Minxia, C. Reference gene selection for real-time quantitative PCR in black medic (Medicago lupulina L.) root tissue under copper stress. J. Agric. Biotechnol. 2014. [Google Scholar] [CrossRef]
- Jiang, X.M.; Zhang, X.Q.; Yan, H.D.; Zhang, Y.; Yang, S.T.; Huang, L.K. Reference gene selection for Real-time quantitative PCR normalization in switchgrass (Panicum virgatum L.) root tissue. J. Agric. Biotechnol. 2014, 22, 55–63. [Google Scholar]
- Xiao, D.; Zhang, N.W.; Zhao, J.J.; Bonnema, G.; Hou, X.L. Validation of reference genes for real-time quantitative PCR normalisation in non-heading Chinese cabbage. Funct. Plant Biol. 2012, 39, 342–350. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Reference gene selection for RT-qPCRnormalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Matos, M.; Correia, S.; Martins, N.; Gonçalves, S.; Romano, A.; Pinto-Carnide, O. Genetic diversity of two endemic and endangered Plantago species. Biochem. Syst. Ecol. 2013, 51, 37–44. [Google Scholar] [CrossRef]
- Xie, F.; Sun, G.; Stiller, J.W.; Zhang, B. Genome-Wide Functional Analysis of the Cotton Transcriptome by Creating an Integrated EST Database. PLoS ONE 2011, 6, e26980. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; Paepe, A.D.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Shriram, V.; Kishor, P.B.K.; Jawali, N.; Shitole, M.G. Enhanced proline accumulation and salt stress tolerance of transgenic indica rice by over-expressing P5CSF129A gene. Plant Biotechnol. Rep. 2010, 4, 37–48. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Greenway, H.; Munns, R. Mechanisms of Salt Tolerance in Nonhalophytes. Annu. Rev. Plant Biol. 2003, 31, 149–190. [Google Scholar] [CrossRef]
- Murgia, I.; Tarantino, D.; Vannini, C.; Bracale, M.; Carravieri, S.; Soave, C. Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to Paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J. 2004, 38, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Mccord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [PubMed]
- Baum, J.A.; Scandalios, J.G. Developmental Expression and Intracellular Localization of Superoxide Dismutases in Maize. Differentiation 1979, 13, 133–140. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Hamalainen, H.K.; Tubman, J.C.; Vikman, S.; Kyrölä, T.; Ylikoski, E.; Warrington, J.A.; Lahesmaa, R. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal. Biochem. 2001, 299, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Paolacci, A.R.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009, 10, 1–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Roche, J.R.; Donaghy, D.J.; Thrush, A.; Sathish, P. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol. Biol. 2010, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, J.E.M. Ruth C, Evaluation of reference genes for quantitative RT-PCR in Lolium temulentum under abiotic stress. Plant Sci. 2009, 176, 390–396. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Chen, W.; Chen, J.; Lu, W.; Lei, C.; Fu, D. Evaluation of New Reference Genes in Papaya for Accurate Transcript Normalization under Different Experimental Conditions. PLoS ONE 2012, 7, e44405. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.R.; Nam, H.Y.; Kim, S.U.; Kim, S.I.; Chang, Y.J. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 2003, 25, 1869–1872. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. MGG 2010, 283, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yan, H.; Jiang, X.; Zhang, Y.; Zhang, X.; Ji, Y.; Zeng, B.; Xu, B.; Yin, G.; Lee, S. Reference gene selection for quantitative real-time reverse-transcriptase PCR in orchardgrass subjected to various abiotic stresses. Gene 2014, 553, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.; Zhang, X.; Wang, J.; Xiao, M.; Zhou, M.; Huang, L.K.; Gang, N.; Wang, P.; Yang, Z.; Ji, L. Transcriptional Profiles of Drought-Related Genes in Modulating Metabolic Processes and Antioxidant Defenses in Lolium multiflorum. Front. Plant Sci. 2016, 7, 519. [Google Scholar] [CrossRef]
- Wang, X.; Ma, X.; Huang, L.; Zhang, X. Identification of the Valid Reference Genes for Quantitative RT-PCR in Annual Ryegrass (Lolium multiflorum) under Salt Stress. Molecules 2015, 20, 4833–4847. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Chen, S.; Liu, Z.; Shan, H.; Luo, H.; Guan, Z.; Chen, F. Reference gene selection for quantitative real-time PCR in Chrysanthemum subjected to biotic and abiotic stress. Mol. Biotechnol. 2011, 49, 192–197. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |



| Rank | Acidic Aluminum Stress | Drought Stress | Heavy Metal Stress | Saline-Alkali Stress | All Samples |
|---|---|---|---|---|---|
| 1 | Unigene71 | Unigene14912 | Unigene71 | Unigene14912 | Unigene14912 |
| (1.48 ± 0.49) | (1.75 ± 0.58) | (0.86 ± 0.29) | (2.43 ± 0.82) | (2.14 ± 0.72) | |
| 2 | eIF4A | Unigene71 | HIS3 | HIS3 | Unigene755 |
| (1.76 ± 0.43) | (1.98 ± 0.71) | (1.56 ± 0.43) | (2.46 ± 0.71) | (2.95 ± 0.93) | |
| 3 | Unigene14912 | eIF4A | Unigene755 | Unigene71 | HIS3 |
| (1.9 ± 0.64) | (2.29 ± 0.55) | (1.56 ± 0.49) | (2.59 ± 0.93) | (3.08 ± 0.87) | |
| 4 | HIS3 | E2 | E2 | SAMDC | eIF4A |
| (2.01 ± 0.56) | (2.68 ± 0.61) | (2.09 ± 0.47) | (2.78 ± 0.78) | (3.35 ± 0.83) | |
| 5 | SAMDC | TBP-1 | Unigene14912 | TBP-1 | Unigene77 |
| (2.41 ± 0.71) | (3.02 ± 0.72) | (2.26 ± 0.77) | (3.45 ± 0.86) | (3.4 ± 1.06) | |
| 6 | Unigene755 | SAMDC | Unigene77 | Unigene755 | Unigene71 |
| (2.51 ± 0.79) | (3.27 ± 0.87) | (2.3 ± 0.71) | (3.59 ± 1.13) | (3.69 ± 1.27) | |
| 7 | Unigene77 | HIS3 | TBP-1 | Unigene77 | TBP-1 |
| (2.63 ± 0.80) | (4.14 ± 1.19) | (2.44 ± 0.57) | (3.96 ± 1.25) | (4.39 ± 1.06) | |
| 8 | 18S rRNA | Unigene755 | eIF4A | eIF4A | E2 |
| (6.46 ± 0.84) | (4.14 ± 1.30) | (2.62 ± 0.65) | (4.11 ± 1.06) | (5.2 ± 1.21) | |
| 9 | TBP-1 | Unigene77 | SAMDC | E2 | SAMDC |
| (8.25 ± 2.03) | (4.23 ± 1.32) | (3.78 ± 1.15) | (5.15 ± 1.25) | (5.22 ± 1.50) | |
| 10 | E2 | 18S rRNA | 18S rRNA | 18S rRNA | 18S rRNA |
| (8.83 ± 2.06) | (8.82 ± 1.17) | (5.09 ± 0.67) | (9.7 ± 1.46) | (8.57 ± 1.17) | |
| 11 | GAPDH | GAPDH | GAPDH | GAPDH | GAPDH |
| (14.04 ± 3.44) | (11.84 ± 2.90) | (14.26 ± 3.31) | (12.12 ± 2.82) | (13.36 ± 3.19) |
| Rank | Acidic Aluminum Stress | Drought Stress | Heavy Metal Stress | Saline-Alkali Stress | All Samples |
|---|---|---|---|---|---|
| 1 | 18S rRNA | eIF4A | Unigene71 | TBP-1 | Unigene14912 |
| (0.545) | (0.273) | (0.26) | (0.445) | (0.899) | |
| 2 | SAMDC | E2 | HIS3 | Unigene14912 | eIF4A |
| (0.843) | (0.594) | (0.374) | (0.764) | (0.928) | |
| 3 | Unigene71 | Unigene14912 | TBP-1 | eIF4A | HIS3 |
| (0.955) | (0.606) | (0.426) | (0.892) | (1.003) | |
| 4 | HIS3 | TBP-1 | eIF4A | Unigene755 | Unigene755 |
| (0.962) | (0.734) | (0.483) | (1.043) | (1.074) | |
| 5 | Unigene14912 | HIS3 | Unigene755 | HIS3 | 18S rRNA |
| (1.092) | (0.874) | (0.511) | (1.195) | (1.162) | |
| 6 | eIF4A | SAMDC | E2 | E2 | Unigene77 |
| (1.207) | (0.891) | (0.550) | (1.232) | (1.318) | |
| 7 | Unigene755 | Unigene71 | 18S rRNA | 18S rRNA | Unigene71 |
| (1.417) | (1.122) | (0.895) | (1.319) | (1.562) | |
| 8 | Unigene77 | Unigene755 | Unigene77 | Unigene77 | TBP-1 |
| (1.425) | (1.129) | (0.976) | (1.474) | (1.684) | |
| 9 | TBP-1 | 18S rRNA | Unigene14912 | SAMDC | E2 |
| (3.224) | (1.327) | (1.015) | (1.487) | (1.803) | |
| 10 | E2 | Unigene77 | SAMDC | Unigene71 | SAMDC |
| (3.319) | (1.472) | (1.604) | (1.693) | (2.062) | |
| 11 | GAPDH | GAPDH | GAPDH | GAPDH | GAPDH |
| (3.707) | (3.174) | (4.223) | (3.064) | (3.564) |
| Method | Stability (High→Low) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| All samples | |||||||||||
| BestKeeper | Unigene14912 | eIF4A | HIS3 | Unigene755 | Unigene77 | TBP-1 | 18S rRNA | E2 | Unigene71 | SAMDC | GAPDH |
| NormFinder | Unigene14912 | eIF4A | HIS3 | Unigene755 | 18S rRNA | Unigene77 | Unigene71 | TBP-1 | E2 | SAMDC | GAPDH |
| GeNorm | Unigene77|Unigene755 | HIS3 | Unigene14912 | eIF4A | 18S rRNA | Unigene71 | TBP-1 | E2 | SAMDC | GAPDH | |
| RefFinder | Unigene14912 | HIS3 | eIF4A | Unigene755 | Unigene77 | 18S rRNA | TBP-1 | Unigene71 | E2 | SAMDC | GAPDH |
| Drought stress | |||||||||||
| BestKeeper | eIF4A | Unigene14912 | E2 | Unigene71 | TBP-1 | SAMDC | 18S rRNA | HIS3 | Unigene755 | Unigene77 | GAPDH |
| NormFinder | eIF4A | E2 | Unigene14912 | TBP-1 | HIS3 | SAMDC | Unigene71 | Unigene755 | 18S rRNA | Unigene77 | GAPDH |
| GeNorm | E2|TBP-1 | eIF4A | Unigene14912 | 18S rRNA | SAMDC | HIS3 | Unigene71 | Unigene755 | Unigene77 | GAPDH | |
| RefFinder | eIF4A | E2 | TBP-1 | Unigene14912 | SAMDC | HIS3 | Unigene71 | 18S rRNA | Unigene755 | Unigene77 | GAPDH |
| Saline-alkali stress | |||||||||||
| BestKeeper | HIS3 | SAMDC | Unigene14912 | TBP-1 | Unigene71 | eIF4A | Unigene755 | Unigene77 | E2 | 18S rRNA | GAPDH |
| NormFinder | TBP-1 | Unigene14912 | eIF4A | Unigene755 | HIS3 | E2 | 18S rRNA | Unigene77 | SAMDC | Unigene71 | GAPDH |
| GeNorm | Unigene755|Unigene14912 | HIS3 | Unigene71 | SAMDC | Unigene77 | TBP-1 | eIF4A | E2 | 18S rRNA | GAPDH | |
| RefFinder | Unigene14912 | TBP-1 | HIS3 | Unigene755 | eIF4A | SAMDC | Unigene71 | E2 | Unigene77 | 18S rRNA | GAPDH |
| Acidic aluminum stress | |||||||||||
| BestKeeper | eIF4A | Unigene71 | HIS3 | Unigene14912 | SAMDC | Unigene755Unigene77 | 18S rRNA | TBP-1 | E2 | GAPDH | |
| NormFinder | 18S rRNA | SAMDC | Unigene71 | HIS3 | Unigene14912 | eIF4A | Unigene755 | Unigene77 | TBP-1 | E2 | GAPDH |
| GeNorm | SAMDC|Unigene14912 | HIS3 | eIF4A | Unigene71 | 18S rRNA | Unigene77 | Unigene755 | TBP-1 | E2 | GAPDH | |
| RefFinder | SAMDC | HIS3 | Unigene14912 | eIF4A | 18S rRNA | Unigene71 | Unigene755 | Unigene77 | TBP-1 | E2 | GAPDH |
| Heavy metal stress | |||||||||||
| BestKeeper | Unigene71 | HIS3 | E2 | Unigene755 | TBP-1 | eIF4A | 18S rRNA | Unigene77 | Unigene14912 | SAMDC | GAPDH |
| NormFinder | Unigene71 | HIS3 | TBP-1 | eIF4A | Unigene755 | E2 | 18S rRNA | Unigene77 | Unigene14912 | SAMDC | GAPDH |
| GeNorm | E2|Unigene71 | HIS3 | Unigene755 | 18S rRNA | Unigene77 | Unigene14912 | eIF4A | TBP-1 | SAMDC | GAPDH | |
| RefFinder | Unigene71 | HIS3 | E2 | Unigene755 | TBP-1 | eIF4A | 18S rRNA | Unigene77 | Unigene14912 | SAMDC | GAPDH |
| Genes Name | Accession ID | Genes Description | Primer Sequence (5′→3′) Forward and Reverse | Tm/°C | Amplicon Length/bp | Amplification Efficiency | R2 |
|---|---|---|---|---|---|---|---|
| 18S rRNA | NC003071.7 | 18S ribosomal RNA | F:GATAGGAAGAGCCGACAT R:ATACGAACCGTGAAAGCG | 58.5 | 156 | 87.1% | 0.999 |
| SAMDC | HO177535 | S-adenosylmethionine decarboxylase | F:TAGCCTGTTCATCTACTC R:AAGAATCCTTGGAATGGT | 55 | 82 | 92.2% | 0.982 |
| GAPDH | GR518033 | Glyceraldehyde-3-phosphate dehydrogenase | F:TCTGACCGTTAGACTTGAGAAGG R:CTTGAGCTTACCCTCAGACTCCT | 59 | 135 | 100.8% | 0.991 |
| TBP-1 | GO924783 | Proteasome regulatory subunit 6A homolog | F:TGCTTAGTTCCCCTAAGATAGTGA R:CTGAGACCAAACACGATTTCA | 60 | 112 | 104.2% | 0.993 |
| eIF4A | GO924770 | Eukaryotic initiation factor4 alpha | F:AACTCAACTTGAAGTGTTGGAGTG R:AGATCTGGTCCTGGAAAGAATATG | 59 | 168 | 95.4% | 0.991 |
| E2 | GO924794 | Ubiquitin-conjugatingenzyme | F:CGGTTCTGTGCCAAAATGT R:CAGCTATCTCCAACGGTTCA | 61.6 | 111 | 92.9% | 0.990 |
| HIS3 | KM454863.1 | Imidazoleglycerol-phosphate dehydratase | F:ACTTCAAGACTGATCTGCGTTTCC R:CATGGATGGCACAAAGGTTGG | 63.3 | 80 | 108% | 0.994 |
| Unigene71 | unknown | F:CTTGGAGGAAGAGGGACGAG R:GAGACAGGAGAGAGTGGCGG | 61.3 | 134 | 93.4% | 0.992 | |
| Unigene77 | unknown | F:CAGGTAAAAGGAACAAACAT R:TCAATTCAGTAATCAAAGGC | 56.9 | 98 | 89.6% | 0.980 | |
| Unigene755 | unknown | F:AAATGATAAACTGACCAAAC R:CCACTCAAGAGCCAAAATAG | 55 | 101 | 91.3% | 0.980 | |
| Unigene14912 | unknown | F:TACACCCATGAGAATGCCAC R:TGTTACCTTGAGGAGTCGGA | 59.4 | 110 | 91.2% | 0.982 | |
| P5CS1 | JX470539 | Delta-1-pyrroline-5-carboxylate | F:ATAACCAATGCTATCCCTGAC R:TCTTAGTCGTTGCCTTGA | 51 | 160 | ||
| Cyt-Cu/Zn SOD | JQ269677 | Cytosolic Cu/Zn superoxide | F:GGCTGAGTATCCCATTT R:CTGCCTTTGCTGTTCT | 57 | 87 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Qi, X.; Yan, H.; Huang, L.; Nie, G.; Zhang, X. Reference Gene Selection for Quantitative Real-Time Reverse-Transcriptase PCR in Annual Ryegrass (Lolium multiflorum) Subjected to Various Abiotic Stresses. Molecules 2018, 23, 172. https://doi.org/10.3390/molecules23010172
Liu Q, Qi X, Yan H, Huang L, Nie G, Zhang X. Reference Gene Selection for Quantitative Real-Time Reverse-Transcriptase PCR in Annual Ryegrass (Lolium multiflorum) Subjected to Various Abiotic Stresses. Molecules. 2018; 23(1):172. https://doi.org/10.3390/molecules23010172
Chicago/Turabian StyleLiu, Qiuxu, Xiao Qi, Haidong Yan, Linkai Huang, Gang Nie, and Xinquan Zhang. 2018. "Reference Gene Selection for Quantitative Real-Time Reverse-Transcriptase PCR in Annual Ryegrass (Lolium multiflorum) Subjected to Various Abiotic Stresses" Molecules 23, no. 1: 172. https://doi.org/10.3390/molecules23010172
APA StyleLiu, Q., Qi, X., Yan, H., Huang, L., Nie, G., & Zhang, X. (2018). Reference Gene Selection for Quantitative Real-Time Reverse-Transcriptase PCR in Annual Ryegrass (Lolium multiflorum) Subjected to Various Abiotic Stresses. Molecules, 23(1), 172. https://doi.org/10.3390/molecules23010172






