Determination of the Main Nucleosides and Nucleobases in Natural and Cultured Ophiocordyceps xuefengensis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Calibration Curves, Limit of Detection (LOD), Limit of Quantification (LOQ) and Method Validation
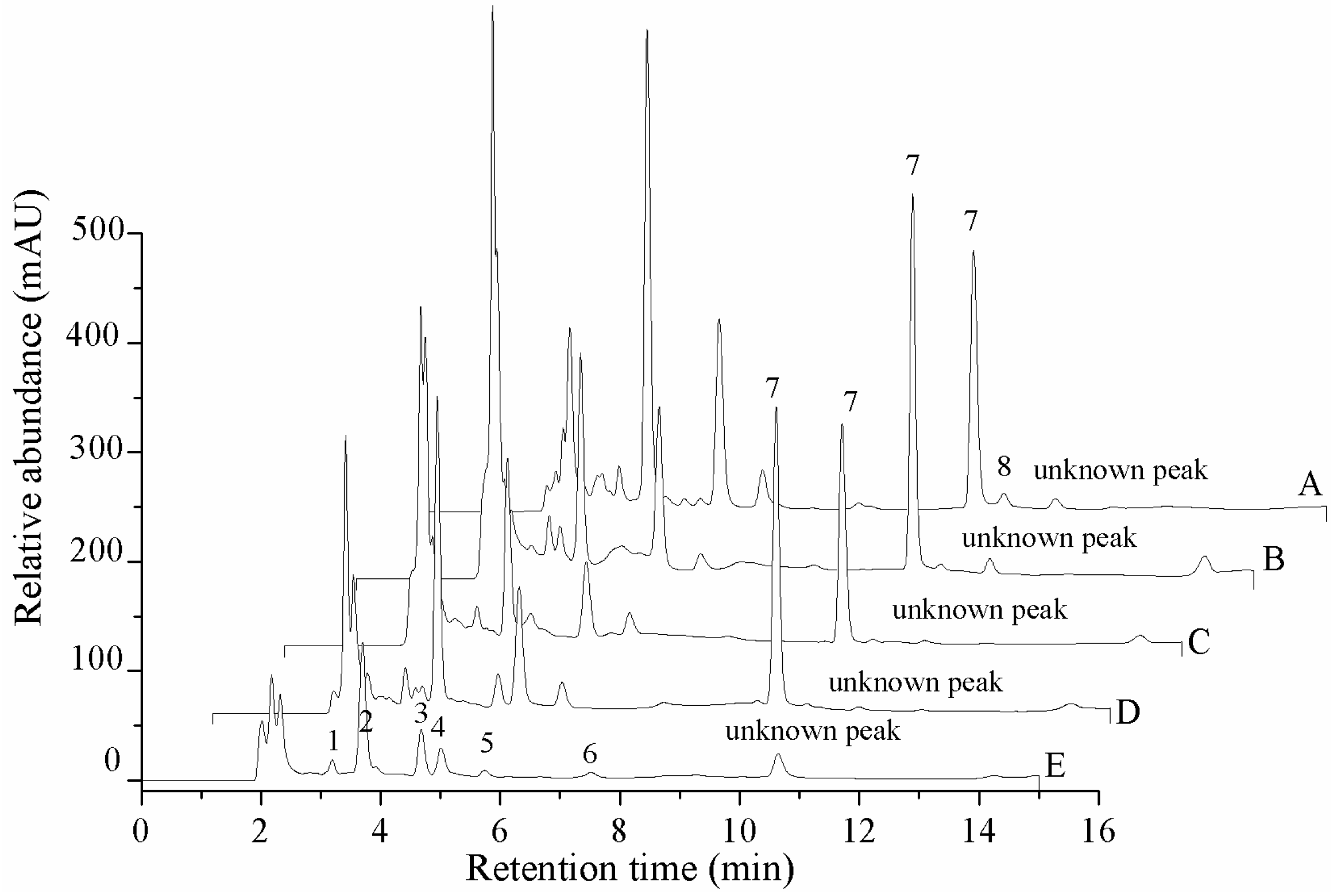
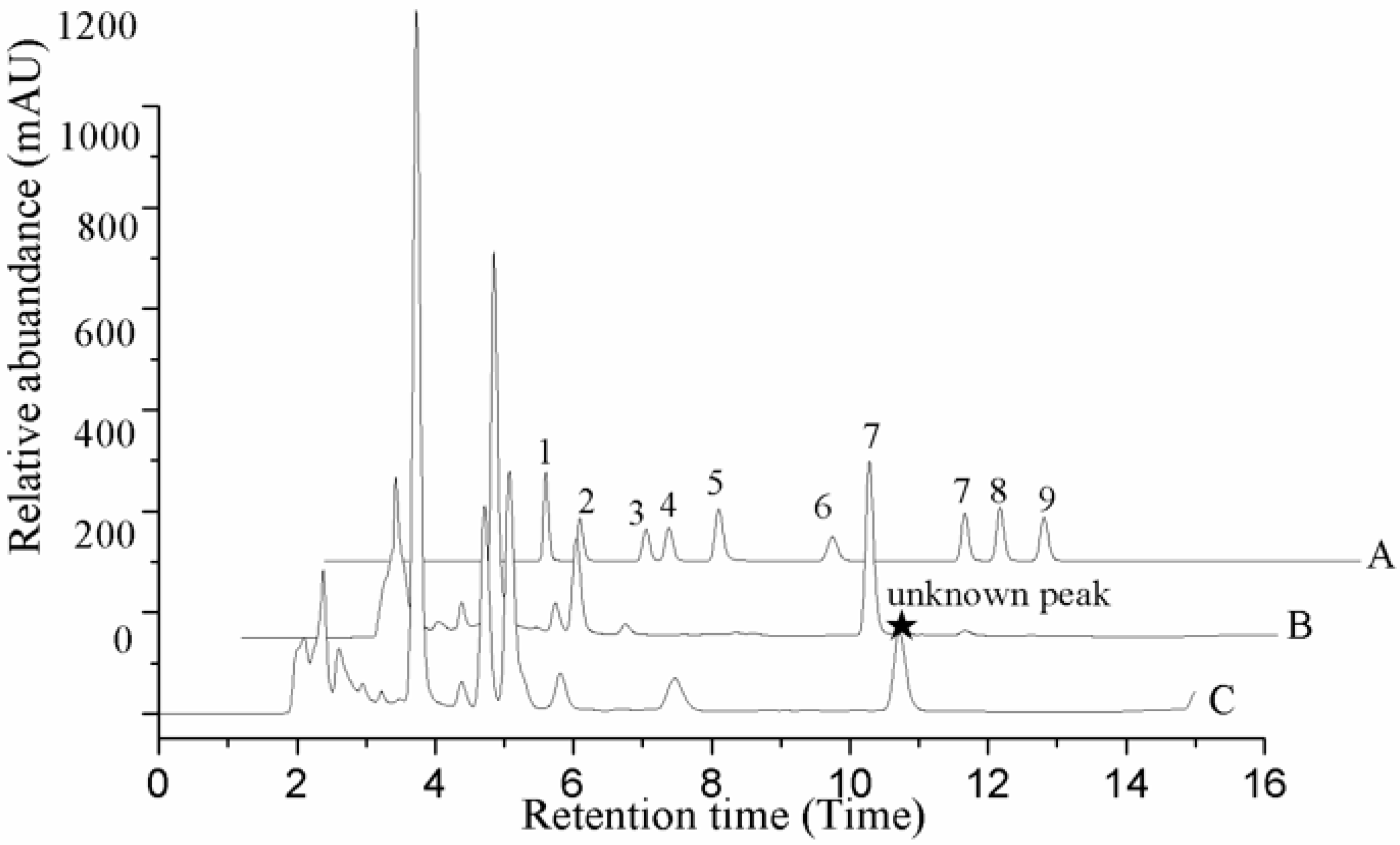
2.2. Contents of the Nucleosides and Nucleobases in Different Tissues
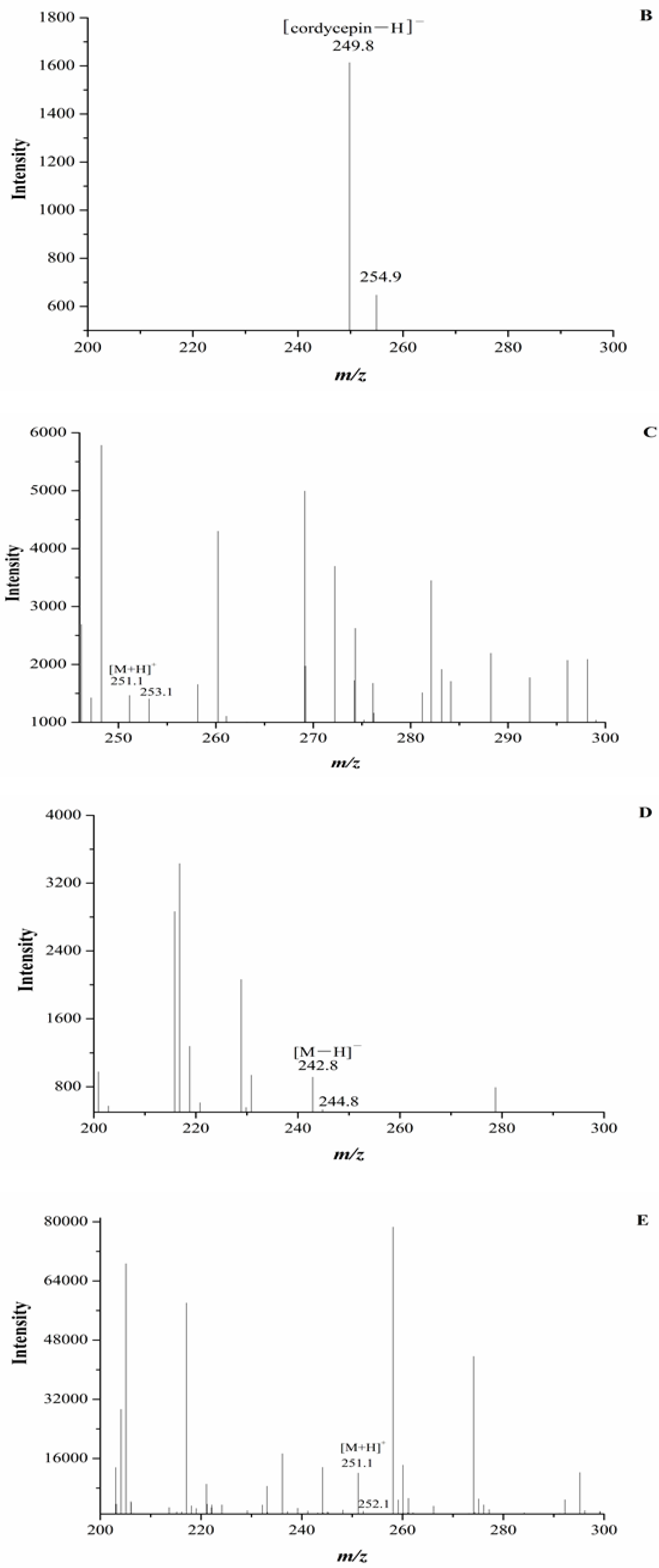
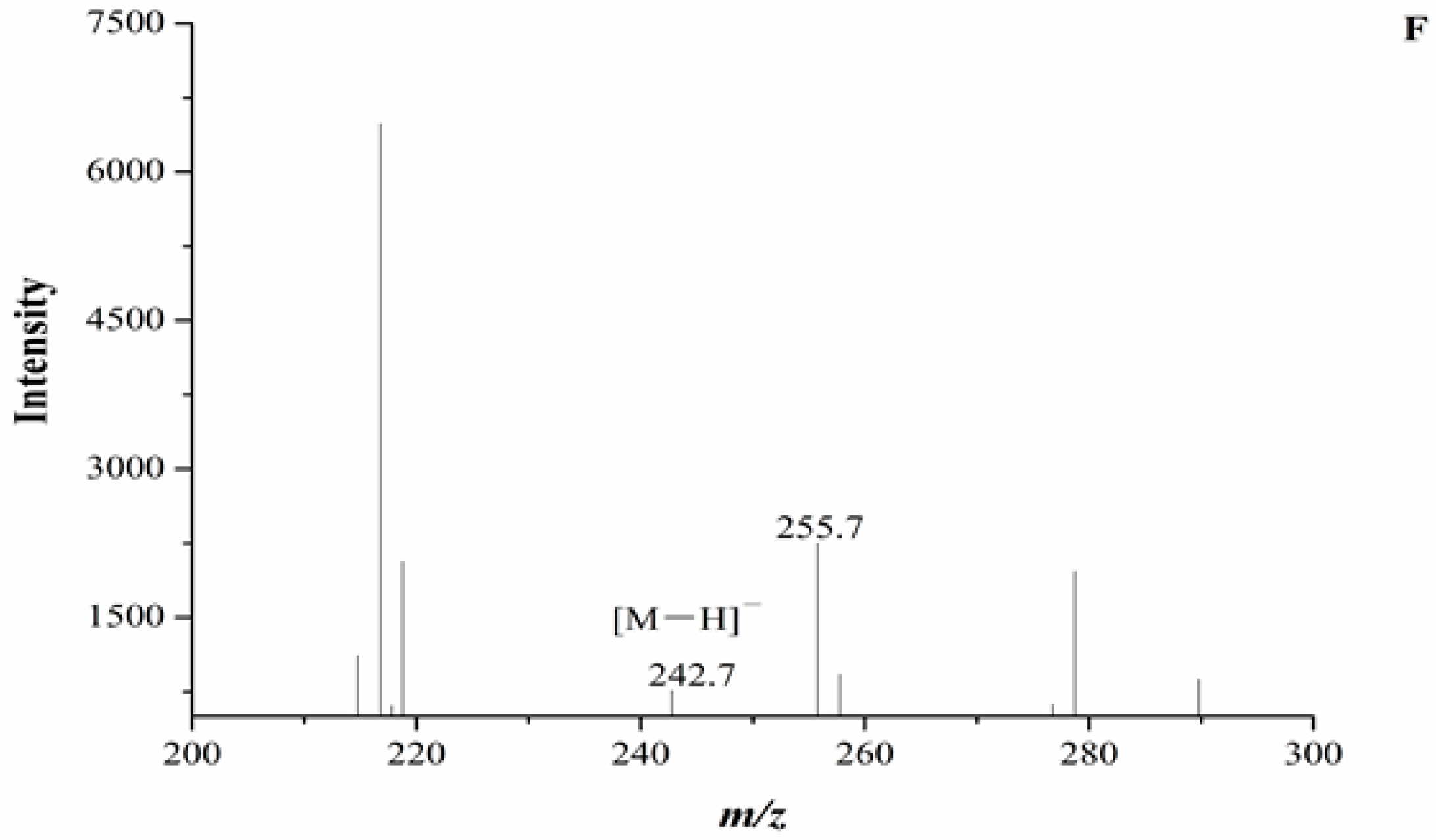
2.3. Identification of Cordycepin
3. Experimental
3.1. Materials
3.2. Inoculum Preparation and Flask Cultures
3.3. Artificial Cultivation of Coremium
3.4. Extraction of the Nucleoside Analogues and Nucleobases
3.5. Standard Samples and the Linear Regression Equation
3.6. HPLC Analysis
3.7. ESI/TOF-MS Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002, 3, 415–424. [Google Scholar] [CrossRef]
- Xiao, J.H.; Qi, Y.; Xiong, Q. Nucleosides, a valuable chemical marker for quality control in traditional Chinese medicine Cordyceps. Recent Pat. Biotechnol. 2013, 7, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Yang, F.Q.; Tsim, K.W. Quality control of Cordyceps sinensis, a valued traditional Chinese medicine. J. Pharm. Biomed. Anal. 2006, 41, 1571–1584. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.X.; Wang, S.; Nie, S.; Marcone, M. Properties of Cordyceps sinensis: A review. J. Funct. Foods 2013, 5, 550–569. [Google Scholar] [CrossRef]
- Tuli, H.S.; Sharma, A.K.; Sandhu, S.S.; Kashyap, D. Cordycepin: A bioactive metabolite with therapeutic potential. Life Sci. 2013, 93, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Luo, L.; Dressel, W.; Shadier, G.; Krumbiegel, D.; Schmidtke, P.; Zepp, F.; Meyer, C.U. Cordycepin is an immunoregulatory active ingredient of Cordyceps sinensis. Am. J. Chin. Med. 2008, 36, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.H.; Lee, C.M.; Lee, S.W.; Seo, S.Y.; Seo, M.J.; Kang, B.W.; Jeong, Y.K.; Choi, Y.J.; Yang, K.M.; Jo, W.S. Cordycepin-enriched Cordyceps militaris induces immunomodulation and tumor growth delay in mouse-derived breast cancer. Oncol. Rep. 2013, 30, 1996–2002. [Google Scholar] [CrossRef] [PubMed]
- Pharmacopoeia Commission of PRC. Pharmacopoeia of the People′s Republic of China, vol. I; Chemical Industry Press: Beijing, China, 2005; p. 75. [Google Scholar]
- Wen, T.C.; Zhu, R.C.; Kang, J.C.; Huang, M.H. Ophiocordyceps xuefengensis sp. nov. from larvae of Phassus nodus (Hepialidae) in Hunan Province, southern China. Phytotaxa 2013, 123, 41–50. [Google Scholar]
- Zheng, B.; Xie, F.Y.; Cai, G.H.; Zhu, R.C.; Li, K.; Gao, S.Q.; Tan, D.B.; Hao, X.Y.; Qin, Y.H. Effects of Ophiocordyceps xuefengensis on proliferation of DC-CIK cells and activity of killing HepG-2 cells by DC-CIK cells. Chin. J. Immunol. 2015, 31, 189–192. [Google Scholar]
- Zou, J.; Wu, L.; He, Z.M.; Zhang, P.; Chen, Z.H. Biological characteristics and cultivation of Ophiocordyceps xuefengensis. Mycosystema 2017, 36, 1104–1110. [Google Scholar]
- Zhao, H.Q.; Wang, X.; Li, H.M.; Yang, B.; Yang, H.J.; Huang, L. Characterization of nucleosides and nucleobases in natural Cordyceps by HILIC-ESI/TOF/MS and HILIC-ESI/MS. Molecules 2013, 18, 9755–9769. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Kan, L.J.; Nie, S.P.; Chen, H.H.; Cui, S.W.; Phillips, A.O.; Phillips, G.O.; Li, Y.J.; Xie, M.Y. A comparison of chemical composition, bioactive components and antioxidant activity of natural and cultured Cordyceps sinensis. LWT-Food Sci. Technol. 2015, 63, 2–7. [Google Scholar] [CrossRef]
- Xie, J.; Huang, L.; Hu, W.; He, Y.; Wong, K. Analysis of the main nucleosides in Cordyceps sinensis by LC/ESI-MS. Molecules 2010, 15, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.Y.; Han, H.; Wang, B.; Li, N.; Dong, T.T.; Zhang, T.; Tsim, K.W. Fast simultaneous determination of 13 nucleosides and nucleobases in Cordyceps sinensis by UHPLC-ESI-MS/MS. Molecules 2015, 20, 21816–21825. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Li, S.P.; Xiang, J.J.; Lai, C.M.; Yang, F.Q.; Gao, J.L.; Wang, Y.T. Qualitative and quantitative determination of nucleosides, bases and their analogues in natural and cultured Cordyceps by pressurized liquid extraction and high performance liquid chromatography–electrospray ionization tandem mass spectrometry HPLC–ESI–MS/MS). Anal. Chim. Acta 2006, 567, 218–228. [Google Scholar]
- Zeng, W.B.; Yu, H.; Ge, F.; Yang, J.Y.; Chen, Z.H.; Wang, Y.B.; Dai, Y.D.; Adams, A. Distribution of nucleosides in populations of Cordyceps cicadae. Molecules 2014, 19, 6123–6141. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.H.; Xiao, D.M.; Sun, Z.H.; Xiong, Q.; Liang, Z.Q.; Zhong, J.J. Chemical compositions and antimicrobial property of three edible and medicinal Cordyceps species. J. Food Agric. Environ. 2009, 7, 91–100. [Google Scholar]
- Xiao, J.H. Cordyceps spp. Fermentation, Its Metabolites Separation and Their Bioactivity Study; East China University of Science and Technology: Shanghai, China, 2010; pp. 83–88. [Google Scholar]
Sample Availability: Samples of the Ophiocordyceps xuefengensis are available from the authors. |
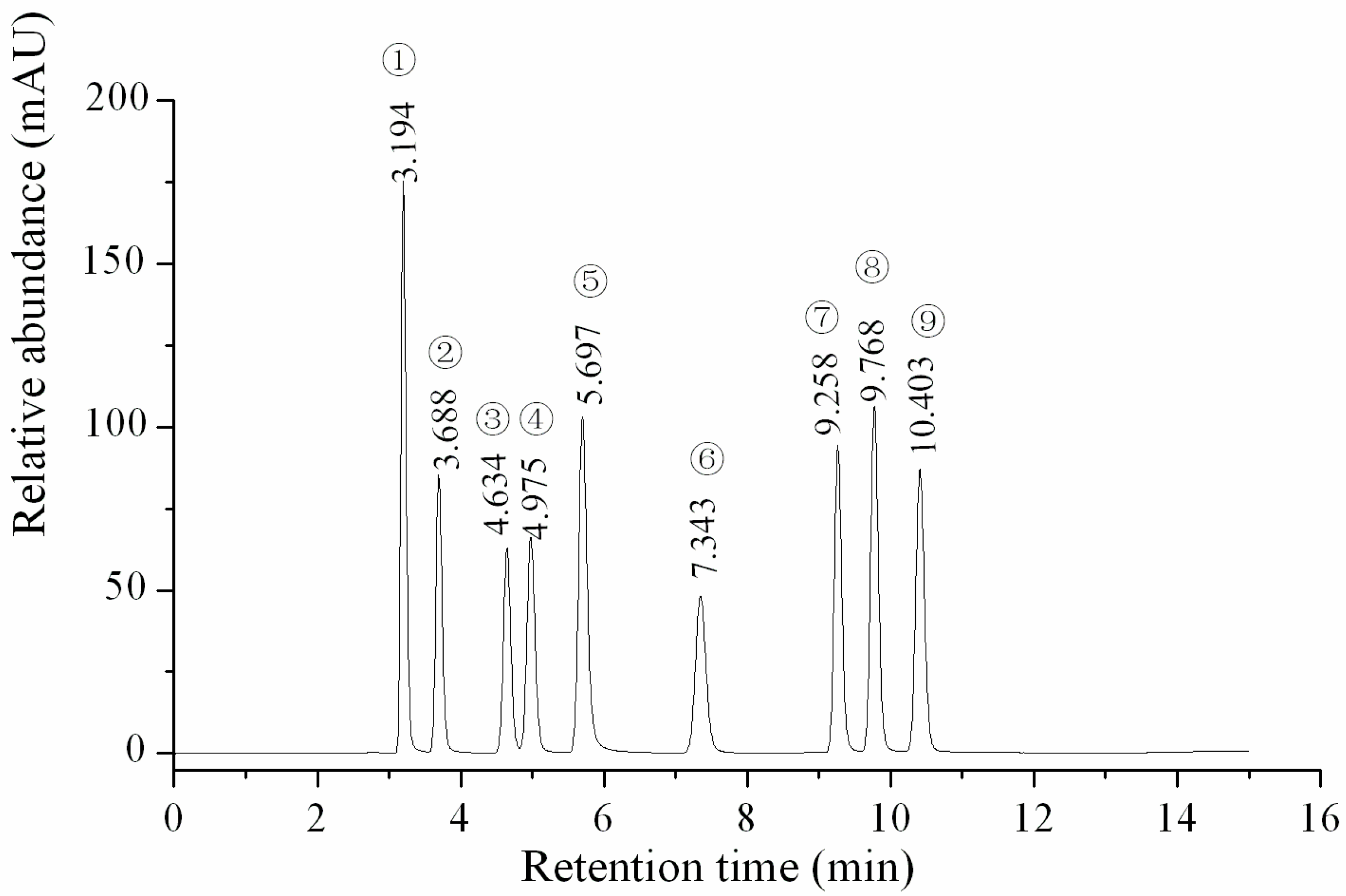


| Analyte | Linear Regression Data | LOD (µg/mL) | LOQ (µg/mL) | ||
|---|---|---|---|---|---|
| Regression Equation | r2 (n = 9) | Linear Range (µg/mL) | |||
| Uracil | Y = 8.54854× 107x + 19748.4 | 0.99986 | 0.5–99.0 | 0.005 | 0.015 |
| Uridine | Y = 5.30058 × 107x − 6067.1 | 0.99997 | 0.5–149.3 | 0.010 | 0.050 |
| Inosine | Y = 4.48713 × 107x + 4348.1 | 0.99995 | 0.5–148.5 | 0.010 | 0.100 |
| Guanosine | Y = 5.31942 × 107x − 37180.3 | 0.99902 | 0.5–147.8 | 0.010 | 0.050 |
| Adenine | Y = 9.81843 × 107x − 41387.9 | 0.99972 | 0.5–146.3 | 0.002 | 0.010 |
| Thymidine | Y = 5.06990 × 107x − 9208.8 | 0.99995 | 0.5–147.8 | 0.015 | 0.100 |
| Adenosine | Y = 6.75044 × 107x + 13235.0 | 0.99992 | 0.5–149.3 | 0.010 | 0.050 |
| 2-deoxyadenosine | Y = 7.93610 × 107x + 19809.4 | 0.99991 | 0.5–151.5 | 0.002 | 0.010 |
| Cordycepin | Y = 7.30463 × 107x − 2626.2 | 0.99991 | 0.5–150.0 | 0.010 | 0.050 |
| Analyte | Precision (RSD, %, n = 6) | Stability (RSD, %, n = 6) | Recovery (%, n = 3) | ||
|---|---|---|---|---|---|
| Intraday | Interday | Mean | RSD (%) | ||
| Uracil | 1.64 | 2.21 | 2.05 | 96.9 | 5.81 |
| Uridine | 1.21 | 1.94 | 1.54 | 100.6 | 4.78 |
| Inosine | 1.00 | 2.29 | 2.25 | 79.4 | 3.68 |
| Guanosine | 2.12 | 1.75 | 1.88 | 80.6 | 4.71 |
| Adenine | 0.96 | 2.32 | 2.29 | 80.6 | 2.93 |
| Thymidine | 1.11 | 1.60 | 1.76 | 102.7 | 1.32 |
| Adenosine | 1.29 | 2.61 | 1.71 | 103.1 | 4.38 |
| 2-deoxyadenosine | 1.68 | 1.49 | 0.59 | 101.5 | 2.75 |
| Cordycepin | 1.64 | 2.80 | 1.63 | 90.4 | 2.88 |
| Analyte | Natural Fruit Body | Cadaver of Phasus Nodus | Cultured Mycelium | Cultured Stroma | Cultured Coremium |
|---|---|---|---|---|---|
| Cordycepin | nd | nd | nd | nd | nd |
| 2-deoxyriboside | nd | nd | 62.8 ± 4.9 | nd | nd |
| Adenosine | 761.5 ± 56.4aA | nd | 623.9 ± 15.4bB | 721.2 ± 31.8aAB | 582.9 ± 42.6bB |
| Thymidine | nd | 66.8 ± 6.6 | nd | nd | nd |
| Adenine | 83.7 ± 8.9cBC | 34.8 ± 0.8dC | 126.1 ± 9.1bB | 155.9 ± 12.2abAB | 186.6 ± 16.6aA |
| Guanosine | 513.6 ± 17.9cC | 179.7 ± 5.4dD | 688.9 ± 14.8aA | 626.1 ± 24.8bB | 502.1 ± 11.5cC |
| Inosine | 177.6 ± 0.9bB | 207.6 ± 2.5aA | nd | nd | nd |
| Uridine | 975.2 ± 41.6bB | 506.9 ± 8.1cC | 1396.6 ± 49.1aA | 869.7 ± 36.0bB | 995.2 ± 85.6bB |
| Uracil | 69.2 ± 2.7cC | 39.4 ± 0.4eE | 87.1 ± 6.9bB | 138.1 ± 4.8aA | 55.9 ± 1.2dD |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Wu, L.; He, Z.-M.; Zhang, P.; Chen, Z.-H. Determination of the Main Nucleosides and Nucleobases in Natural and Cultured Ophiocordyceps xuefengensis. Molecules 2017, 22, 1530. https://doi.org/10.3390/molecules22091530
Zou J, Wu L, He Z-M, Zhang P, Chen Z-H. Determination of the Main Nucleosides and Nucleobases in Natural and Cultured Ophiocordyceps xuefengensis. Molecules. 2017; 22(9):1530. https://doi.org/10.3390/molecules22091530
Chicago/Turabian StyleZou, Juan, Ling Wu, Zheng-Mi He, Ping Zhang, and Zuo-Hong Chen. 2017. "Determination of the Main Nucleosides and Nucleobases in Natural and Cultured Ophiocordyceps xuefengensis" Molecules 22, no. 9: 1530. https://doi.org/10.3390/molecules22091530




