Multi-Dimensional Spectrum-Effect Relationship of the Impact of Chinese Herbal Formula Lichong Shengsui Yin on Ovarian Cancer
Abstract
:1. Introduction
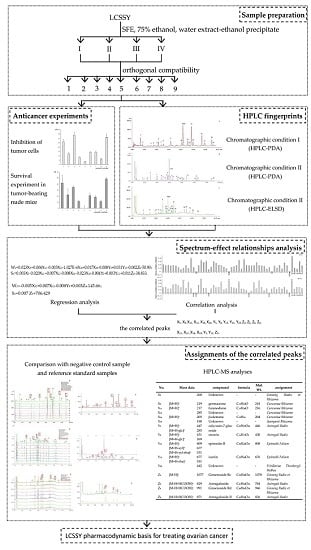
2. Results and Discussion
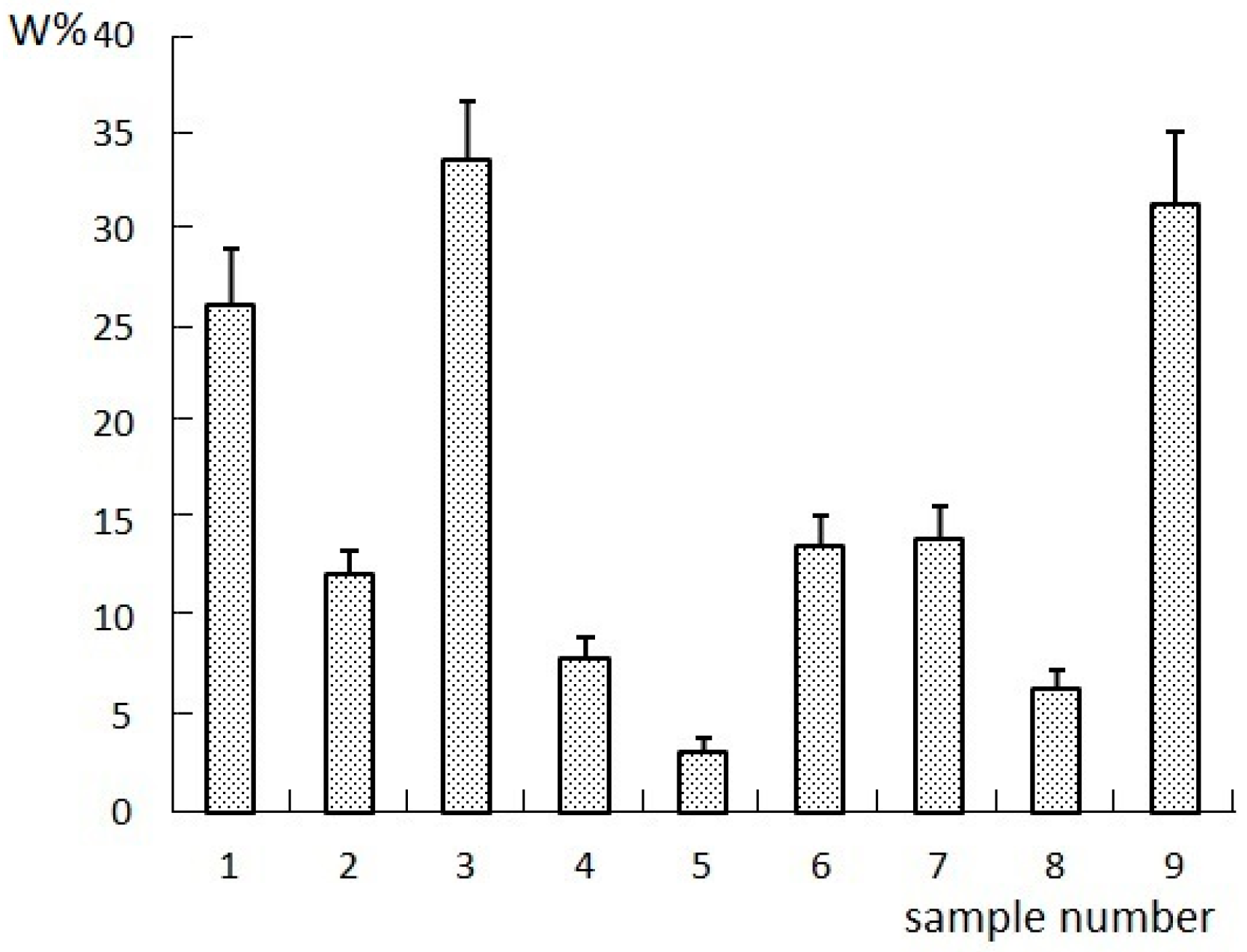
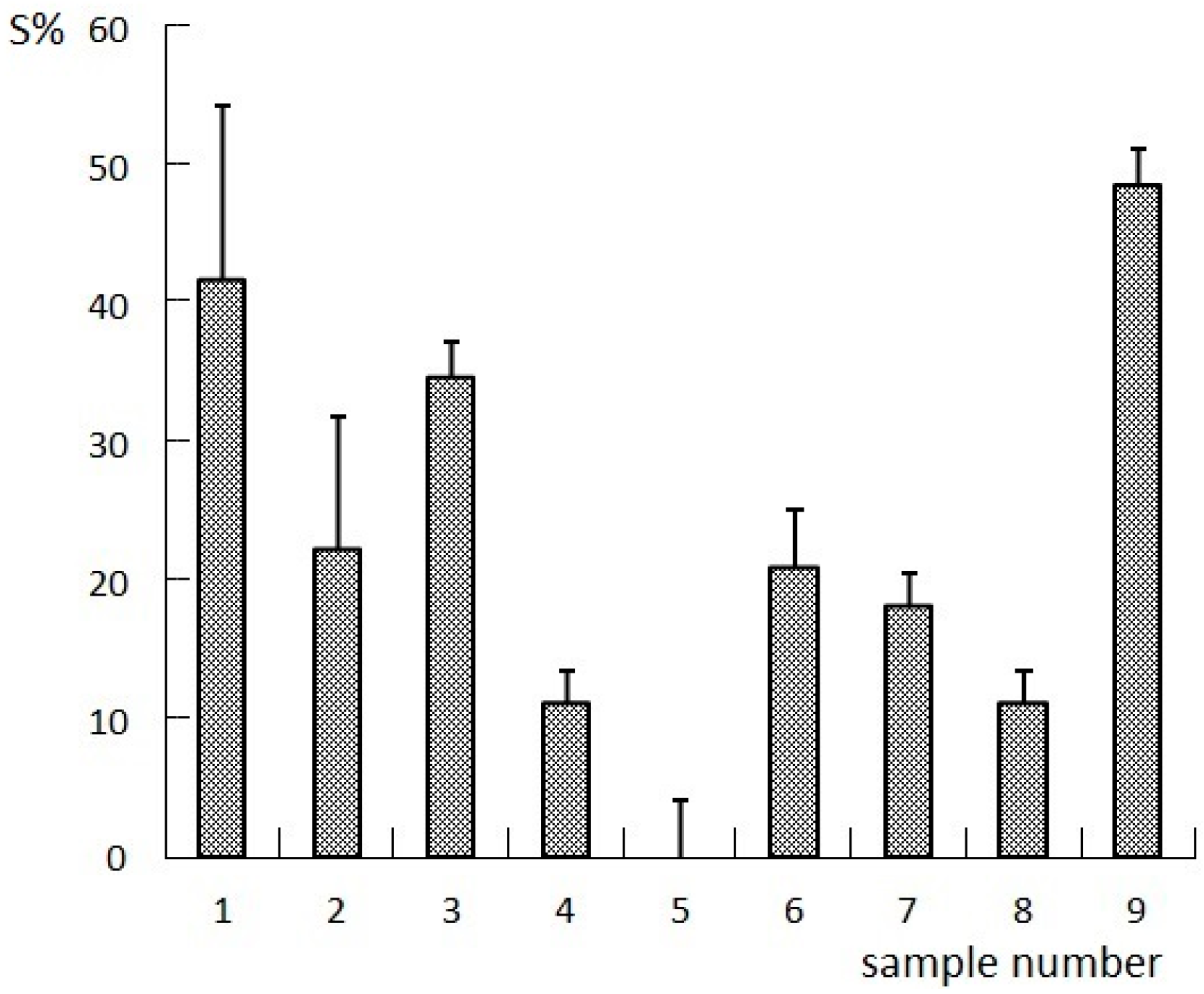
2.1. Results of the Orthogonal Compatibility Anticancer Experiment
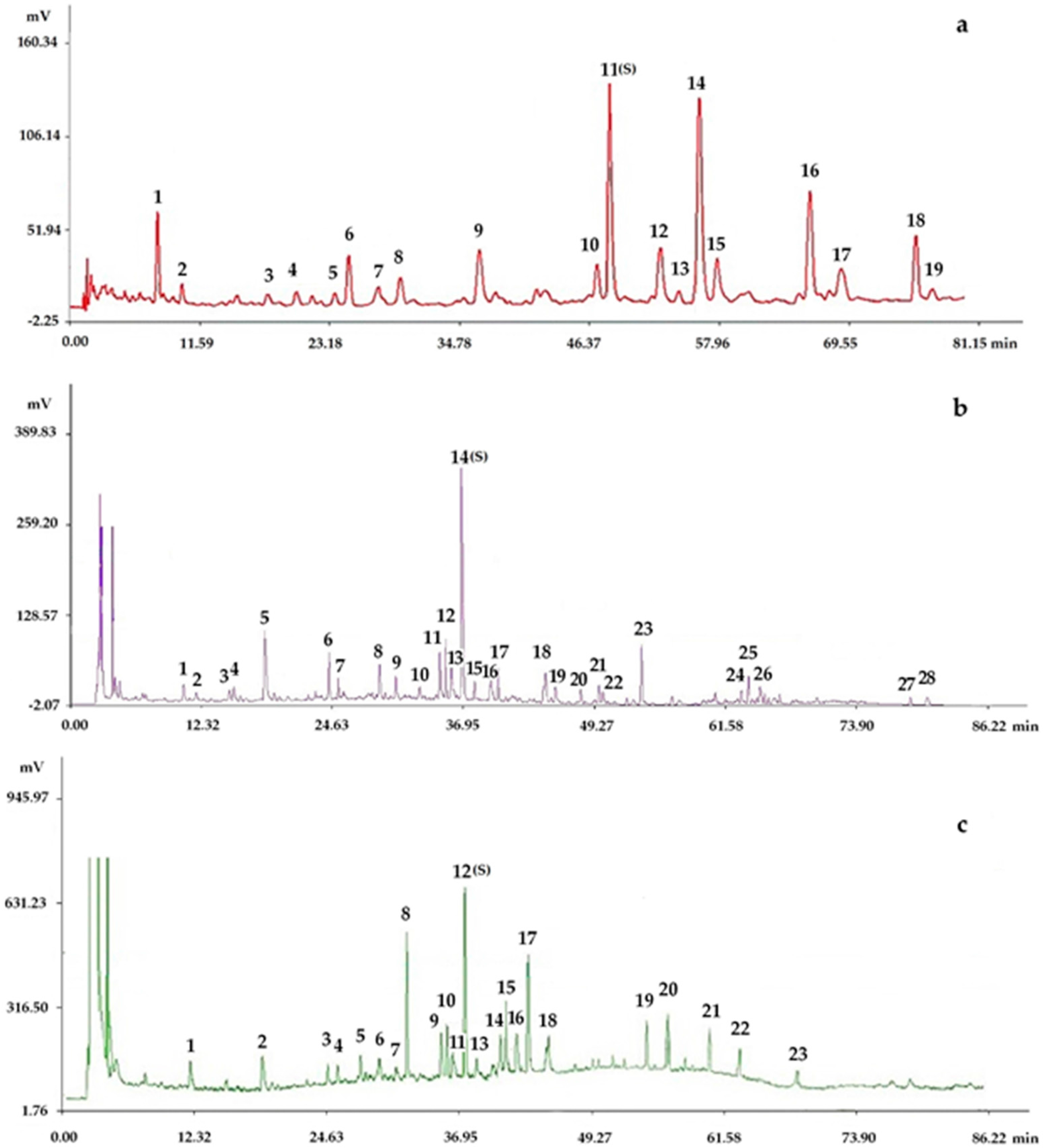
2.2. Results of the HPLC Experiment
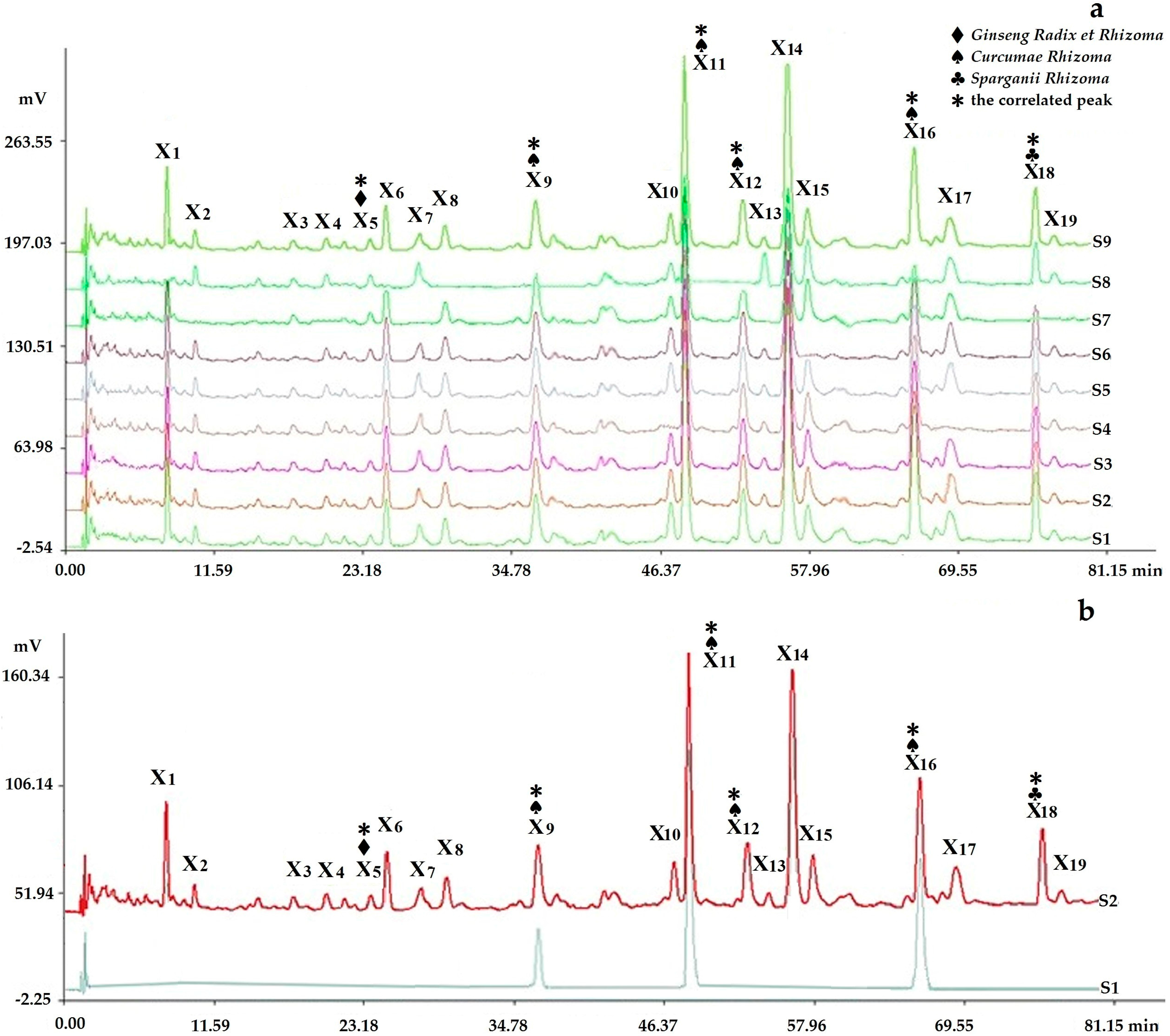
2.2.1. Fingerprints Analyzed by Chromatographic Condition I (HPLC-PDA)
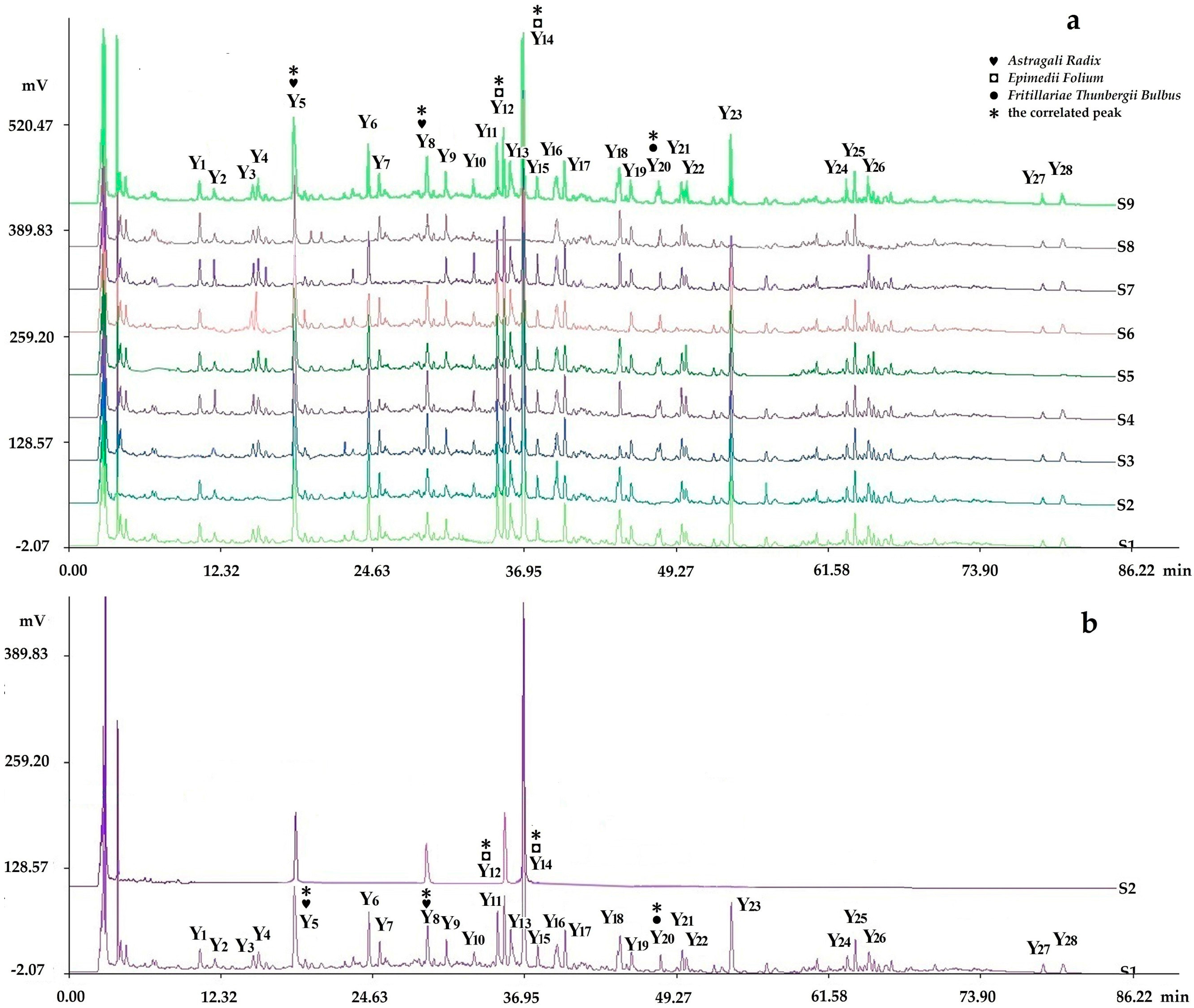
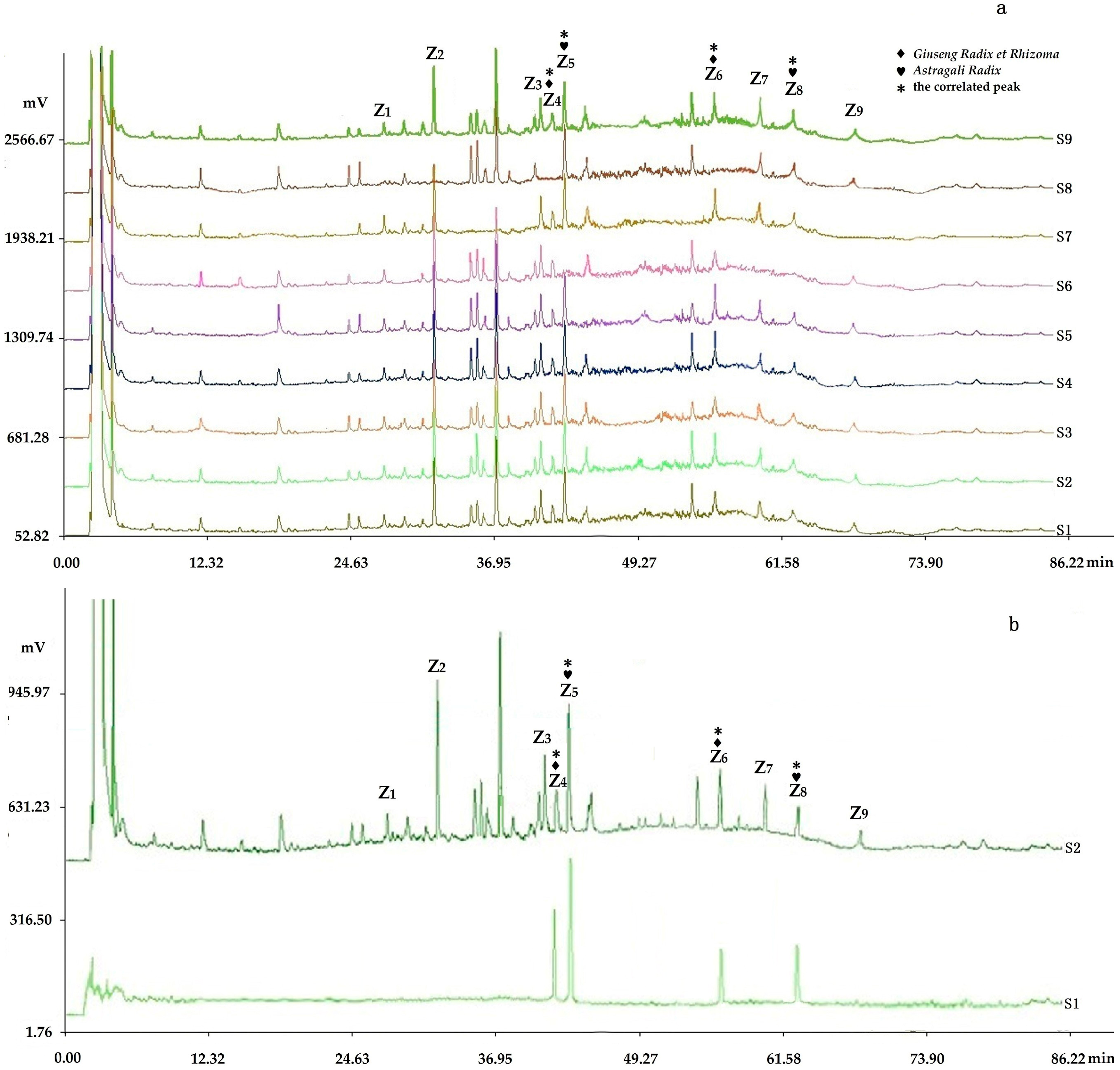
2.2.2. Fingerprints Analyzed by Chromatographic Condition II (HPLC-PDA-ELSD)
2.2.3. Determination of Fingerprints
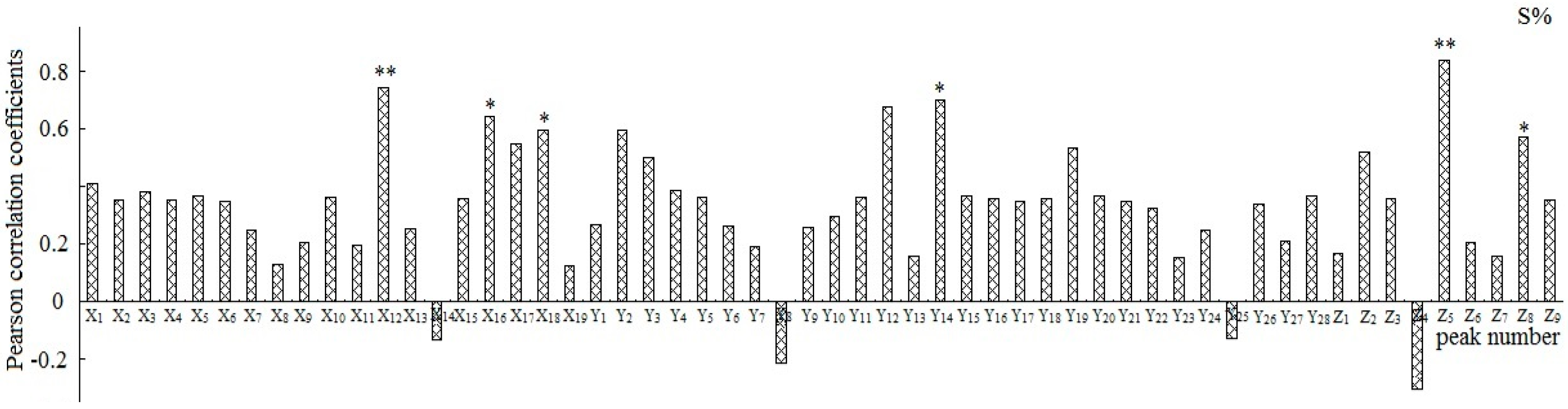
2.3. The Analysis of Spectrum-Effect Relationships
2.3.1. Regression Analysis
ENTER Method
S1 = 0.003X5 − 0.020X11 + 0.007X12 − 0.008X16 − 0.023X18 − 0.004Y5 + 0.003Y12 + 0.011Z6 − 38.833.
STEPWISE Method
S2 = 0.007 Z5 + 706.429.
2.3.2. Correlation Analysis
2.3.3. Integration of the Analytical Results
2.3.4. Assignments of the Correlated Peaks
3. Materials and Methods
3.1. Reagents and Materials
3.2. Sample Preparation
3.3. Anticancer Experiments
3.3.1. Inhibition of Tumor Cells
3.3.2. Survival Experiment in Tumor-Bearing Nude Mice
3.4. Analysis of HPLC Fingerprints
3.4.1. Preparation of Sample Solutions
3.4.2. HPLC Conditions
3.4.3. Validation of Methodology
3.4.4. Determination of Fingerprints
3.5. Analysis of Spectrum-Effect Relationships
3.5.1. Regression Analysis
3.5.2. Correlation Analysis
3.5.3. Assignments of the Correlated Peaks
Comparison of the Fingerprints with Negative Control Samples and Reference Standard Samples
HPLC-MS Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LCSSY | Lichong Shengsui Yin |
| TCMs | Traditional Chinese medicines |
| HPLC-PDA | High-performance liquid chromatography-photodiode array detector |
| HPLC-PDA-ELSD | High-performance liquid chromatography-photodiode array detector -Evaporative Light-scattering Detector |
| HPLC-MS | High-performance liquid chromatography-mass spectrometry detector |
| HPTLC | High performance thin layer chromatography |
| GC-MS | Gas chromatography-mass spectrometry |
| IR | Infrared spectroscopy |
| HPCE | High performance capillary electrophoresis |
| R.S.D. | Relative standard deviation |
| MTT | 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide |
| i.p. | Intraperitoneal injection |
References
- Zhang, L.; Yan, J.B.; Liu, X.M.; Ye, Z.G.; Yang, X.H.; Meyboome, R.; Chan, K.; Shaw, D.; Duez, P. Pharmacovigilance practice and risk control of Traditional Chinese Medicine drugs in China: Current status and future perspective. J. Ethnopharmacol. 2012, 140, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Li, S.; Tan, H.Y.; Wang, N.; Tsao, S.W.; Feng, Y. Current Status of Herbal Medicines in Chronic Liver Disease Therapy: The Biological Effects, Molecular Targets and Future Prospects. Int. J. Mol. Sci. 2015, 16, 28705–28745. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yin, Q.W.; Zhang, A.H.; Sun, H.; Wu, X.H.; Wang, X.J. Preliminary identification of the absorbed bioactive compositions and metabolites in rat plasma after oral administration of Shaoyao-Gancao decoction by ultra-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Pharmacogn. Mag. 2014, 10, 497–502. [Google Scholar]
- Ouyang, E.H.; Zhang, C.G.; Li, X.M. Determination of 5-hydroxymethyl-2-furaldehyde of crude and processed Fructus Corni in freely moving rats using in vivo microdialysis sampling and liquid chromatography. J. Pharmacogn. Mag. 2011, 7, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.H.; Li, Z.G.; Tian, G.H.; Gao, K.; Li, Z.Y.; Zhao, B.S.; Wang, J.; Luo, L.T.; Pan, Q.; Zhang, W.T.; et al. Effects of traditional Chinese medicine on rats with Type II diabetes induced by high-fat diet and streptozotocin: A urine metabonomic study. J. Afr. Health Sci. 2013, 13, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.L.; Xie, M.; Yang, X.Y.; Song, Y.; Yan, C.; Yang, Y.; Zhang, X.; Liu, Z.Z.; Tian, Y.X.; Wang, Y.; et al. Spectrum-Effect Relationships as a Systematic Approach to Traditional Chinese Medicine Research: Current Status and Future Perspectives. Molecules 2014, 19, 17897–17925. [Google Scholar] [CrossRef] [PubMed]
- Pei, K.; Cai, H.; Duan, Y.; Qiao, F.X.; Tu, S.C.; Liu, X.; Wang, X.L.; Song, X.Q.; Fan, K.L.; Cai, B.C. Evaluation of the Influence of Sulfur-Fumigated Paeoniae Radix Alba on the Quality of Si Wu Tang by Chromatographic and Chemometric Analysis. J. Anal. Methods Chem. 2016, 2016, 8358609. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.M.; Yu, J.Y.; Ding, M.J.; Zhao, M.; Xue, X.Y.; Che, C.T.; Wang, S.M.; Zhao, B.; Meng, J. Liquid Chromatography-diode Array Detector-electrospray Mass Spectrometry and Principal Components Analyses of Raw and Processed Moutan Cortex. J. Pharmacogn. Mag. 2016, 12, 50–56. [Google Scholar] [PubMed]
- Karthika, K.; Paulsamy, S. TLC and HPTLC Fingerprints of Various Secondary Metabolites in the Stem of the Traditional Medicinal Climber, Solena amplexicaulis. J. Indian J. Pharm. Sci. 2015, 77, 111–116. [Google Scholar] [PubMed]
- Singh, M.; Tamboli, E.T.; Kamal, Y.T.; Ahmad, W.; Ansari, S.H.; Ahmad, S. Quality control and in vitro antioxidant potential of Coriandrum sativum Linn. J. Pharm. Bioallied Sci. 2015, 7, 280–283. [Google Scholar] [PubMed]
- Guo, Y.; Lv, B.; Wang, J.; Liu, Y.; Sun, S.; Xiao, Y.; Lu, L.; Xiang, L.; Yang, Y.; Qu, L.; et al. Analysis of Chuanxiong Rhizoma and its active components by Fourier transform infrared spectroscopy combined with two-dimensional correlation infrared spectroscopy. J. Spectrochim. Acta Mol. Biomol. Spectrosc. 2016, 15, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.F.; Cheung, H.Y. Development of capillary electrophoresis fingerprint for quality control of rhizoma Smilacis Glabrae. J. Phytochem. Anal. 2011, 22, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Konieczyński, P. Electrochemical fingerprint studies of selected medicinal plants rich in flavonoids. J. Acta Pol. Pharm. 2015, 72, 655–661. [Google Scholar]
- Li, A.P.; Li, Z.Y.; Sun, H.F.; Li, K.; Qin, X.M.; Du, G.H. Comparison of Two Different Astragali Radix by a 1H-NMR-Based Metabolomic Approach. J. Proteome Res. 2015, 14, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Yang, W.; Guo, D.; Wu, J.; Zhang, J.; Qiu, S.; Yao, C.; Cui, Y.; Wu, W. Selective ion monitoring of quinochalcone C-glycoside markers for the simultaneous identification of Carthamus tinctorius L. in eleven Chinese patent medicines by UHPLC/QTOF MS. J. Pharm. Biomed. Anal. 2016, 117, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.S.; Lin, Z.J.; Xiao, M.L.; Niu, H.J.; Zhang, B. The spectrum-effect relationship-a rational approach to screening effective compounds, reflecting the internal quality of Chinese herbal medicine. J. Chin. J. Nat. Med. 2016, 14, 177–184. [Google Scholar] [CrossRef]
- Han, F.J.; Wang, Y.H.; Wang, X.X.; Sui, L.H. Effect of compound prescription Lichong Shengsui Yin on the morphology of ovarian tumor in rats induced by dimethylbenzanthracine. Acta Chin. Med. Pharm. 2006, 34, 31–33. [Google Scholar]
- Han, F.J.; Wang, Y.H.; Sui, L.H.; Wang, X.X.; Liu, J. Effect of compound prescription Lichong Shengsui Yin drug serum on the colony formation and expression of Bcl-2 protein in ovarian cancer. Chin. J. Tradit. Med. Sci. Technol. 2005, 12, 345–346. [Google Scholar]
- Han, F.J.; Xu, F.; Wang, Y.H.; Sui, L.H.; Wang, X.X. Effect of compound prescription Lichong Shengsui Yin on the expression of P53 and PCNA in the ovarian tumor tissue of rats. Chin. J. Tradit. Med. Sci. Technol. 2007, 14, 402–403. [Google Scholar]
- Gong, L.H.; Han, F.J.; Wang, X.X.; Hou, L.H.; Wu, X.K.; Fan, M.M. Effect of Lichong Shengsui Yin on gene expression of ovarian cancer cell line SKOV3. J. Shandong Univ. Tradit. Chin. Med. 2007, 31, 334–337. [Google Scholar]
- Huai, Q.J.; Han, F.J.; Wang, X.X.; Qiu, L.N.; Wu, X.K. Effect of traditional Chinese medicine compound of Lichong Shengsui Yin on expressions of Casp-8 gene and CXCL2 gene in ovarian cancer cellline SKOV3. Matern. Child Health Care China 2010, 25, 1689–1692. [Google Scholar]
- Qiu, L.N.; Han, F.J.; Wu, X.K.; Hou, L.H. Anti-tumor effect of Lichong Shengsui Yin on ovarian tum or tissue and the impact on VEGF expression in Fischer 344 Rats. World J. Intergr. Tradit. West. Med. 2010, 5, 939–941. [Google Scholar]
- Han, F.J.; Sui, L.H.; Ma, R.; Wang, X.X. Clinical study on the effect of Lichong Shengsui Yin on immune state of the patients with ovarian epithelial ovarian cancer. Inf. Tradit. Chin. Med. 2003, 20, 37–38. [Google Scholar]
- Liu, L.; Lu, Y.; Shao, Q.; Cheng, Y.Y.; Qu, H.B. Binary chromatographic fingerprinting for quality evaluation of Radix Ophiopogonis by high-performance liquid chromatography coupled with ultraviolet and evaporative light-scattering detectors. J. Sep. Sci. 2007, 30, 2628–2637. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.Z. Study on the quality of Rhizoma Curcumae. Acta Pharm. Sin. 2011, 36, 849–853. [Google Scholar]
- Ma, E.; Wang, X.; Li, Y.; Sun, X.; Tai, W.; Li, T.; Guo, T. Induction of apoptosis by furanodiene in HL60 leukemia cells through activation of TNFR1 signaling pathway. Cancer Lett. 2008, 18, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yang, F.Q.; Li, S.P.; Lao, S.C.; Conceição, E.L.; Fung, K.P.; Wangl, Y.T.; Lee, S.M. Furanodiene induces G2/M cell cycle arrest and apoptosis through MAPK signaling and mitochondria-caspase pathway in human hepatocellular carcinoma cells. Cancer Biol. Ther. 2007, 6, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, G.; Zhao, J.; Ding, H.; Cunningham, C.; Chen, F.; Flynn, D.C.; Reed, E.; Li, Q.Q. Antiproliferative effect of beta-elemene in chemoresistant ovarian carcinoma cells is mediated through arrest of the cell cycle at the G2-M phase. Cell. Mol. Life Sci. 2005, 62, 894. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.Q.; Ding, X.; Jia, Y. Anti-tumor effect of beta-elemene in glioblastoma cells depends on p38 MAPK activation. Cancer Lett. 2008, 264, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Wang, G.; Zhang, M.; Cuff, C.F.; Huang, L.; Reed, E. β-Elemene, a novel plant-derived antineoplastic agent, increases cisplatin chemosensitivity of lung tumor cells by triggering apoptosis. Oncol. Rep. 2009, 22, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.S.; Li, Q.D.; Zou, B.B. In vitro combination characterization of the new anticancer plant drug β-elemene with taxanes against human lung carcinoma. Int. J. Oncol. 2007, 31, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zheng, Y.; Que, Z.; Zhang, L.; Lin, S.; Le, V.; Liu, J.; Tian, J. Astragaloside IV inhibits progression of lung cancer by mediating immune function of Tregs and CTLs by interfering with IDO. J. Cancer Res. Clin. Oncol. 2014, 140, 1883. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.D.; Gu, J.F.; Zhang, M.H.; Yuan, J.R.; Zhao, B.J.; Jiang, J.; Jia, X.B. Astragaloside IV inhibits migration and invasion in human lung cancer A549 cells via regulating PKC-α-ERK1/2-NF-κB pathway. Int. Immunol. 2014, 23, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Xu, D.; Xia, Q.; Wang, P.; Rong, C.; Su, Y. Reversal of p-glycoprotein-mediated multidrug resistance of human hepatic cancer cells by astragaloside II. J. Pharm. Pharmacol. 2012, 64, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.G.; Sun, H.X.; Ye, Y.P. Ginsenoside Rd from Panax notoginseng is cytotoxic towards HeLa cancer cells and induces apoptosis. Chem. Biodivers. 2006, 3, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J. Involvement of melastatin type transient receptor potential 7 channels in ginsenoside Rd-induced apoptosis in gastric and breast cancer cells. J. Ginseng Res. 2013, 37, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Jin, Y.R.; Lim, W.C.; Ji, S.M.; Cho, J.Y.; Ban, J.J.; Lee, S.K. Ginsenoside Rc and Re stimulate c-fos expression in mcf-7 human breast carcinoma cells. Arch. Pharma. Res. 2003, 26, 53. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, X.; Meng, J.; Wang, Z.Y. An anticancer agent icaritin induces sustained activation of the extracellular signal-regulated hinase (ERK) pathway and inhibits growth of breast cancer cells. Eur. J. Pharmacol. 2011, 658, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Dong, P.; Wang, J.; Zhang, J.; Gu, J.; Wu, X.; Wu, W.; Fei, X.; Zhang, Z.; Wang, Y.; et al. Icariin, a natural flavonol glycoside, induces apoptosis in human hepatoma SMMC-7721 cells via a ROS/JNK-dependent mitochondrial pathway. Cancer Lett. 2010, 298, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, H.; Zhu, M.; Oub, Y.W.; Zhang, J.; Luo, H.S.; Luo, R.Y.; Wu, J.Z.; Mao, M.; Liu, X.H.; et al. Icariin exterts negative effects on human gastric cancer cell invasion and migration by vasodilator-stimulated phosphoprotein via Racl pathway. Eur. J. Pharmacol. 2010, 635, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Sun, J.; Hu, S.Q.; Liu, J. Icariin Induced B16 Melanoma Tumor Cells Apoptosis, Suppressed Tumor Growth and Metastasis. Iran J. Public Health 2014, 43, 847–848. [Google Scholar] [PubMed]
- Li, J.W.; Wang, S.Z.; Zhao, F.J. Icariin inhibits proliferation, migration and invasion of SKOV3 ovarian cancer cells through upregulation of miR-519d. Prog. Anat. Sci. 2015, 21, 471–474. [Google Scholar]
- Cui, J.L.; Guo, S.X.; Xiao, P.G. Antitumor and antimicrobial activities of endophytic fungi from medicinal parts of Aquilaria sinensis. J. Zhejiang Univ. Sci. B 2011, 12, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, H.; Lu, Y.; Tan, B.J.; Xu, L.; Lawal, T.O.; Mahady, G.B.; Liu, D. Menoprogen, a TCM Herbal Formula for Menopause, Increases Endogenous E2 in an Aged Rat Model of Menopause by Reducing Ovarian Granulosa Cell Apoptosis. J. Biomed. Res. Int. 2016, 2016, 2574637. [Google Scholar]
- Kannan, N.; Sakthivel, K.M.; Guruvayoorappan, C. Anti-tumor and Chemoprotective Effect of Bauhinia tomentosa by Regulating Growth Factors and Inflammatory Mediators. J. Asian Pac. J. Cancer Prev. 2015, 16, 8119–8126. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.F.; Zhou, X.; Shi, Z.N.; Li, Y.M.; Li, Z.C. Study on spectrum-effect relationship of rhizoma Rhei, cortex Magnoliae Officinalis, fructus Aurantii Immaturus and their formula. J. Chromatogr. Sci. 2013, 51, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Wang, X.B.; Luo, J.G.; Kong, L.Y. Seasonal Variation of Alkaloid Contents and Anti-Inflammatory Activity of Rhizoma coptidis Based on Fingerprints Combined with Chemometrics Methods. J. Chromatogr. Sci. 2015, 53, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kao, C.P.; Wu, K.C.; Liao, C.R.; Ho, Y.L.; Chang, Y.S. Chemical compositions, chromatographic fingerprints and antioxidant activities of Andrographis Herba. J. Mol. 2014, 19, 18332–18350. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of Germacrone, furanodiene, β-elemene, calycosin-7-glucoside, ononin, epimedin B, icariin, astragalosideare available from the authors. |

| No. | Mass Data | Compound | Formula | Mol. Wt. | Assignment | |
|---|---|---|---|---|---|---|
| X5 | 268 | Unknown | - | - | Ginseng Radix et Rhizoma | |
| X9 | [M + H]+ | 219 | germacrone | C15H22O | 218 | Curcumae Rhizome |
| X11 | [M + H]+ | 217 | furanodiene | C15H20O | 216 | Curcumae Rhizome |
| X12 | 285 | Unknown | - | - | Curcumae Rhizome | |
| X16 | [M + H]+ | 205 | β-elemene | C15H24 | 204 | Curcumae Rhizome |
| X18 | 180 | Unknown | - | - | Sparganii Rhizoma | |
| Y5 | [M + H]+ | 447 | calycosin-7-glucoside | C22H22O10 | 446 | Astragali Radix |
| [M + H − glc]+ | 285 | |||||
| Y8 | [M + H]+ | 431 | ononin | C16H13O4 | 430 | Astragali Radix |
| [M + H − glc]+ | 269 | |||||
| Y12 | [M + H]+ | 809 | epimedin B | C38H48O19 | 808 | Epimedii Folium |
| [M + H − xyl]+ | 677 | |||||
| [M + H – xyl − rha]+ | 531 | |||||
| Y14 | [M + H]+ | 677 | icariin | C33H40O15 | 676 | Epimedii Folium |
| [M + H − rha]+ | 531 | |||||
| Y20 | 442 | Unknown | - | - | Fritillariae Thunbergii Bulbus | |
| Z4 | [M − H]− | 1077 | Ginsenoside Rc | C53H90O22 | 1078 | Ginseng Radix et Rhizoma |
| Z5 | [M – H + HCOOH]− | 829 | Astragaloside | C41H68O14 | 784 | Astragali Radix |
| Z6 | [M – H + HCOOH]− | 991 | Ginsenoside Rd | C48H82O18 | 946 | Ginseng Radix et Rhizoma |
| Z8 | [M – H + HCOOH]− | 871 | Astragaloside II | C43H70O15 | 826 | Astragali Radix |
| Factor Level | Composition I(g) A | Composition II(g) B | Composition III(g) C | Composition IV(g) D |
|---|---|---|---|---|
| 1 | 1(6.60) | 1(22.46) | 1(2.70) | 1(8.33) |
| 2 | 0 | 0 | 0 | 0 |
| 3 | 2(13.20) | 2(44.92) | 2(5.40) | 2(16.66) |
| Test | A(g) | B(g) | C(g) | D(g) |
|---|---|---|---|---|
| 1 | 1(6.60) | 1(22.46) | 1(2.70) | 1(8.33) |
| 2 | 1(6.60) | 0 | 0 | 0 |
| 3 | 1(6.60) | 2(44.92) | 2(5.40) | 2(16.66) |
| 4 | 0 | 1(22.46) | 0 | 2(16.66) |
| 5 | 0 | 0 | 2(5.40) | 1(8.33) |
| 6 | 0 | 2(44.92) | 1(2.70) | 0 |
| 7 | 2(13.20) | 1(22.46) | 2(5.40) | 0 |
| 8 | 2(13.20) | 0 | 1(2.70) | 2(16.66) |
| 9 | 2(13.20) | 2(44.92) | 0 | 1(8.33) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Y.; Zhang, Y.; Feng, G.; Yang, Z.; Guan, Q.; Wang, R.; Han, F. Multi-Dimensional Spectrum-Effect Relationship of the Impact of Chinese Herbal Formula Lichong Shengsui Yin on Ovarian Cancer. Molecules 2017, 22, 979. https://doi.org/10.3390/molecules22060979
Wang Y, Li Y, Zhang Y, Feng G, Yang Z, Guan Q, Wang R, Han F. Multi-Dimensional Spectrum-Effect Relationship of the Impact of Chinese Herbal Formula Lichong Shengsui Yin on Ovarian Cancer. Molecules. 2017; 22(6):979. https://doi.org/10.3390/molecules22060979
Chicago/Turabian StyleWang, Yanhong, Yang Li, Yan Zhang, Guan Feng, Zhixin Yang, Qingxia Guan, Rui Wang, and Fengjuan Han. 2017. "Multi-Dimensional Spectrum-Effect Relationship of the Impact of Chinese Herbal Formula Lichong Shengsui Yin on Ovarian Cancer" Molecules 22, no. 6: 979. https://doi.org/10.3390/molecules22060979