Expression of Genes Related to Phenylpropanoid Biosynthesis in Different Organs of Ixeris dentata var. albiflora
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Sequence Analysis of Phenylpropanoid Biosynthetic Genes from I. dentata
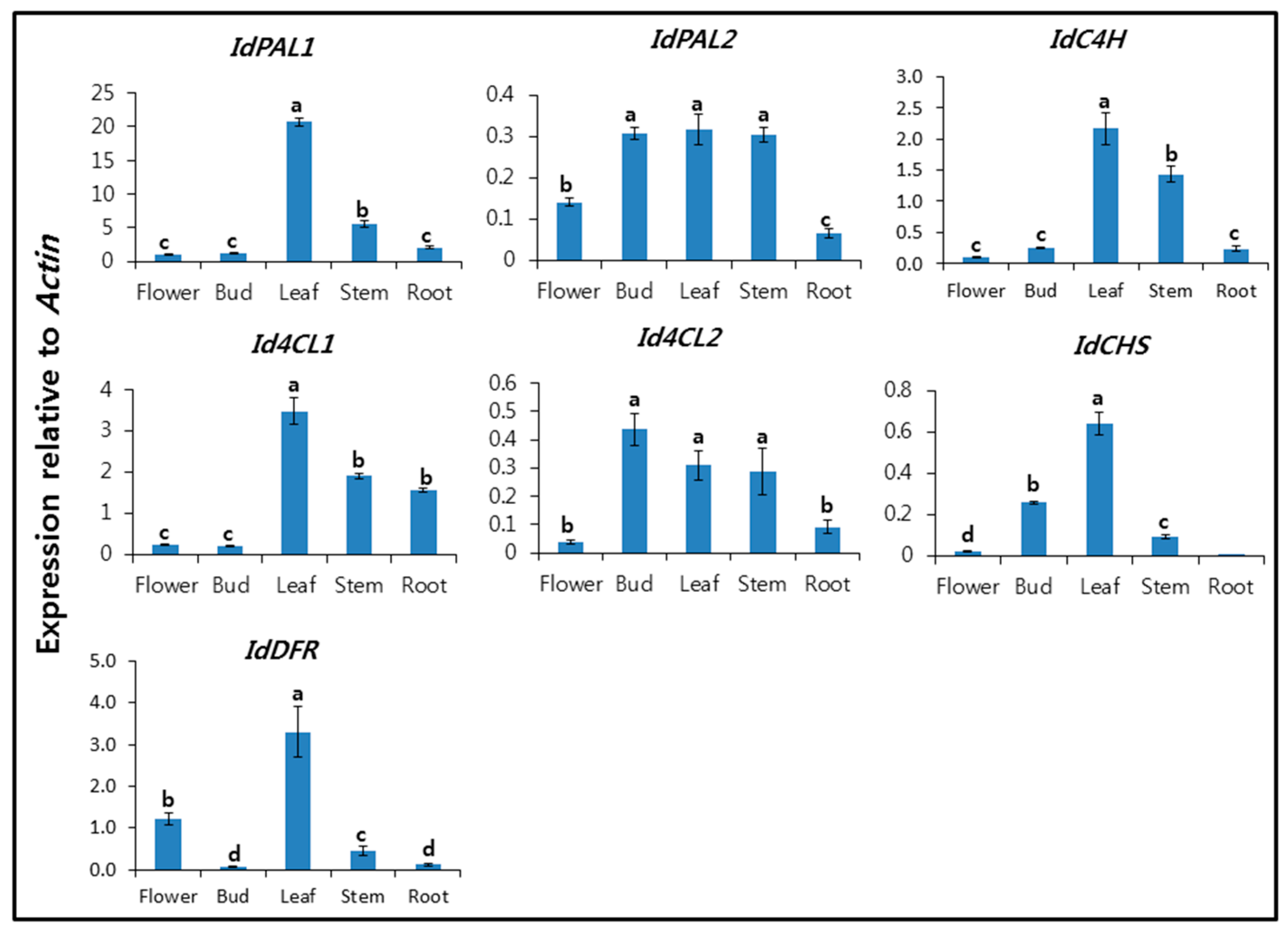
2.2. Expression Profiles of IdPALs, IdC4H, Id4CLs, IdCHS, and IdDFR in Different Organs
2.3. Analysis of Phenylpropanoids in Different Organs of I. dentata
3. Experimental Section
3.1. Plant Materials
3.2. Total RNA Extraction and Quantitative Real Time-PCR (qRT-PCR)
3.3. Sequence Analysis
3.4. Extraction and Analysis of Phenylpropanoids from I. dentata var. albiflora
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ahn, E.M.; Bang, M.H.; Song, M.C.; Park, M.H.; Kim, H.Y.; Kwon, B.M.; Baek, N.I. Cytotoxic and ACAT-inhibitory sesquiterpene lactones from the root of Ixeris dentata forma albiflora. Arch. Pharm. Res. 2006, 29, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Remesy, C.; Jimenez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [PubMed]
- Scalbert, A.; Johnson, I.T.; Saltmarsh, M. Polyphenols: Antioxidants and beyond. Am. J. Clin. Nutr. 2005, 81, 215–217. [Google Scholar]
- Farah, A.; Donangelo, C. Phenolic compounds in coffee. Braz. J. Plant Physiol. 2006, 18, 23–36. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Vrhovsek, U.; Masuero, D.; Gasperotti, M.; Franceschi, P.; Caputi, L.; Viola, R.; Mattivi, F. A versatile targeted metabolomics method for the rapid quantification of multiple classes of phenolics in fruits and beverages. J. Agric. Food Chem. 2012, 60, 8831–8840. [Google Scholar] [CrossRef] [PubMed]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Rozema, J.; Björn, L.O.; Bornman, J.F.; Gaberscik, A.; Häder, D.P.L.; Trost, T.; Germ, M.; Klisch, M.; Gröniger, A.; Sinha, R.P.; et al. The role of UV-B radiation in aquatic and terrestrial ecosystems—An experimental and functional analysis of the evolution of UV-absorbing compounds. J. Photochem. Photobiol. B 2002, 66, 2–12. [Google Scholar] [CrossRef]
- Cheynier, V. Phenolic compounds: From plants to foods. Phytochem. Rev. 2012, 11, 153–177. [Google Scholar] [CrossRef]
- Zhong, C.; Hu, D.; Hou, L.B.; Song, L.Y.; Zhang, Y.J.; Xie, Y.; Tian, L.W. Phenolic compounds from the rhizomes of Smilax china L. and their anti-inflammatory activity. Molecules 2017, 22, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Martens, S.; Teeri, T.; Forkmann, G. Heterologous expression of dihydroflavonol 4-reductases from various plants. FEBS Lett. 2002, 531, 453–458. [Google Scholar] [CrossRef]
- Strack, D.; Wray, V. Methods in Plant Biochemistry; Academic Press: London, UK, 1989; Volume 1, pp. 325–356. [Google Scholar]
- Li, X.; Park, N.I.; Xu, H.; Woo, S.-H.; Park, C.H.; Park, S.U. Differential expression of flavonoid biosynthesis genes and accumulation of phenolic compounds in common buckwheat (Fagopyrum esculentum). J. Agric. Food Chem. 2010, 58, 12176–12181. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Park, C.H.; Li, X.; Kim, Y.B.; Yang, J.; Sung, G.B.; Park, N.I.; Kim, S.; Park, S.U. Accumulation of rutin and betulinic acid and expression of phenylpropanoid and triterpenoid biosynthetic genes in Mulberry (Morus alba L.). J. Agric. Food Chem. 2015, 63, 8622–8630. [Google Scholar]
- Park, N.I.; Xu, H.; Li, X.; Jang, I.H.; Park, S.U. Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus). J. Agric. Food Chem. 2011, 59, 6034–6039. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Kim, Y.B.; Li, X.; Choi, S.R.; Park, S.; Park, J.S.; Lim, Y.P.; Park, S.U. Accumulation of phenylpropanoids by white, blue, and red light irradiation and their organ-specific distribution in Chinese cabbage (Brassica rapa ssp pekinensis). J. Agric. Food Chem. 2015, 63, 6772–6778. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Retey, J. Serine-202 is the putative precursor of the active site dehydroalanine of phenylalanine ammonia lyase. Site-directed mutagenesis studies on the enzyme from parsley (Petroselinum crispum L.). FEBS Lett. 1994, 349, 252–254. [Google Scholar] [CrossRef]
- Stuible, H.P.; Kombrink, E. Identification of the substrate specificity-conferring amino acid residues of 4-coumarate: Coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J. Biol. Chem. 2001, 276, 26893–26897. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Hovel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.P. The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, Y.; Zhan, Y.; Li, Y.; Kawabata, S. Chalcone synthase family genes have redundant roles in anthocyanin biosynthesis and in response to blue/UV-A light in turnip (Brassica rapa; Brassicaceae). Am. J. Bot. 2013, 100, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kim, J.K.; Park, S.-Y.; Zhao, S.; Kim, Y.B.; Lee, S.; Park, S.U. Comparative analysis of flavonoids and polar metabolite profiling of tanno-original and tanno-high rutin buckwheat. J. Agric. Food Chem. 2014, 62, 2701–2708. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Park, N.I.; Li, X.; Xu, H.; Kim, H.M.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase and cinnamate 4-hydroxylase in the phenylpropanoid biosynthesis pathway in garlic (Allium sativum). J. Agric. Food Chem. 2010, 58, 10911–10917. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.K.; Lee, S.Y.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Shin, Y.J.; Tuan, P.A.; Li, X.; Park, Y.; Park, N.I.; Park, S.U. Molecular cloning and characterization of genes involved in rosmarinic acid biosynthesis from Prunella vulgaris. Biol. Pharm. Bull. 2014, 37, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, X.; Cho, D.H.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.U. Accumulation of kaempferitrin and expression of phenylpropanoid biosynthetic genes in kenaf (Hibiscus cannabinus). Molecules 2014, 19, 16987–16997. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Tuan, P.A.; Li, X.; Kim, Y.B.; Kim, H.; Park, C.G.; Yang, J.; Li, C.H.; Park, S.U. Identification of phenylpropanoid biosynthetic genes and phenylpropanoid accumulation by transcriptome analysis of Lycium chinense. BMC Genom. 2013, 14, 802. [Google Scholar] [CrossRef] [PubMed]
- Richet, N.; Tozo, K.; Afif, D.; Banvoy, J.; Legay, S.; Dizengremel, P.; Cabane, M. The response to daylight or continuous ozone of phenylpropanoid and lignin biosynthesis pathways in poplar differs between leaves and wood. Planta 2012, 236, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Park, W.T.; Arasu, M.V.; Al-Dhabi, N.A.; Yeo, S.K.; Jeon, J.; Park, J.S.; Lee, S.Y.; Park, S.U. Yeast extract and silver nitrate induce the expression of phenylpropanoid biosynthetic genes and induce the accumulation of rosmarinic acid in Agastache rugosa cell culture. Molecules 2016, 21, 426. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Thwe, A.A.; Kim, Y.; Li, X.; Cho, J.W.; Park, P.B.; Arasu, M.V.; Al-Dhabi, N.A.; Kim, S.J.; Suzuki, T.; et al. Transcripts of anthocyanidin reductase and leucoanthocyanidin reductase and measurement of catechin and epicatechin in tartary buckwheat. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Kalinova, J.; Vrchotova, N. Level of catechin, myricetin, quercetin and isoquercitrin in buckwheat (Fagopyrum esculentum Moench), changes of their levels during vegetation and their effect on the growth of selected weeds. J. Agric. Food Chem. 2009, 57, 2719–2725. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kim, Y.B.; Kim, Y.; Zhao, S.; Kim, H.H.; Chung, E.; Lee, J.H.; Park, S.U. Differential stress-response expression of two flavonol synthase genes and accumulation of flavonols in tartary buckwheat. J. Plant Physiol. 2013, 170, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Park, W.T.; Xu, H.; Park, N.I.; Park, S.U. Accumulation of tilianin and rosmarinic acid and expression of phenylpropanoid biosynthetic genes in Agastache rugosa. J. Agric. Food Chem. 2012, 60, 5945–5951. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Singh, K.; Sood, P.; Kumar, S.; Ahuja, P. p-Coumarate: CoA ligase as a key gene in the yield of catechins in tea [Camellia sinensis, L., O. KUNTZE]. Funct. Integr. Genom. 2009, 9, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kumar, S.; Rani, A.; Gulati, A.; Ahuja, P. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct. Integr. Genom. 2009, 9, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Cai, R.; Cheng, S.; Du, H.; Wang, Y.; Cheng, S. Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr. J. Biotechnol. 2008, 7, 721–729. [Google Scholar]
- Park, J.H.; Park, N.I.; Xu, H.; Park, S.U. Cloning and characterization of phenylalanine ammonia-lyase and cinnamate 4-hydroxylase and pyranocoumarin biosynthesis in Angelica gigas. J. Nat. Prod. 2010, 73, 1394–1397. [Google Scholar] [CrossRef] [PubMed]
- Tähtiharju, S.; Rijpkema, A.S.; Vetterli, A.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Evolution and diversification of the CYC/TB1 gene family in Asteraceae—A comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol. Biol. Evol. 2012, 29, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 7, 287. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds analyzed herein are not available from the authors because they were isolated on a small scale. They are readily analyzed using the procedures described. |


| Primer Name | Sequence (5′ → 3′) | GenBank Access No. | Size (bp) |
|---|---|---|---|
| IdPAL1(F) | AGCTCCGGCAAGTTCTGGTC | KU724084 | 100 |
| IdPAL1(R) | TCGTCTTCGAATGTGGCGAT | ||
| IdPAL2(F) | CGCCTGGAGAGGAGTTCGAT | KU724085 | 103 |
| IdPAL2(R) | GGAAGTGGAGTCCCATCCCA | ||
| IdC4H(F) | ACCCAGGGCGTTGAGTTCGG | MF034078 | 114 |
| IdC4H(R) | TCGCATCTTCCGCCAGTGCT | ||
| Id4CL1(F) | ACAATCGCGAACAACGAGGA | KU724086 | 129 |
| Id4CL1(R) | CTTCGCATTCGGGAACTTCA | ||
| Id4CL2(F) | CGTCGCACAACAAGTCGATG | KU724087 | 103 |
| Id4CL2(R) | CATCAGCACCACCGAAGCTC | ||
| IdCHS(F) | CCAAGCTCCTCGGTCTTCGT | KU724088 | 104 |
| IdCHS(R) | CTCAGCAAGGTCCTTCGCAA | ||
| IdDFR(F) | ATTGGCGGAAAGGGCAGCAT | MF034079 | 113 |
| IdDFR(R) | GGCTTGGAGGGAATGAGGGAGT | ||
| IdActin(F) | CAATGGAACCGGAATGGTCA | KY419710 | 100 |
| IdActin(R) | CATTACGCCCGTGTGTCGAG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-H.; Park, Y.-J.; Park, S.U.; Lee, S.-W.; Kim, S.-C.; Jung, C.-S.; Jang, J.-K.; Hur, Y.; Kim, Y.B. Expression of Genes Related to Phenylpropanoid Biosynthesis in Different Organs of Ixeris dentata var. albiflora. Molecules 2017, 22, 901. https://doi.org/10.3390/molecules22060901
Lee S-H, Park Y-J, Park SU, Lee S-W, Kim S-C, Jung C-S, Jang J-K, Hur Y, Kim YB. Expression of Genes Related to Phenylpropanoid Biosynthesis in Different Organs of Ixeris dentata var. albiflora. Molecules. 2017; 22(6):901. https://doi.org/10.3390/molecules22060901
Chicago/Turabian StyleLee, Sang-Hoon, Yun-Ji Park, Sang Un Park, Sang-Won Lee, Seong-Cheol Kim, Chan-Sik Jung, Jae-Ki Jang, Yoonkang Hur, and Yeon Bok Kim. 2017. "Expression of Genes Related to Phenylpropanoid Biosynthesis in Different Organs of Ixeris dentata var. albiflora" Molecules 22, no. 6: 901. https://doi.org/10.3390/molecules22060901
APA StyleLee, S.-H., Park, Y.-J., Park, S. U., Lee, S.-W., Kim, S.-C., Jung, C.-S., Jang, J.-K., Hur, Y., & Kim, Y. B. (2017). Expression of Genes Related to Phenylpropanoid Biosynthesis in Different Organs of Ixeris dentata var. albiflora. Molecules, 22(6), 901. https://doi.org/10.3390/molecules22060901






