Anti-Helicobacter pylori Activity of Isocoumarin Paepalantine: Morphological and Molecular Docking Analysis
Abstract
:1. Introduction
2. Results
2.1. Anti-H. pylori Activity
2.2. Computational Methods
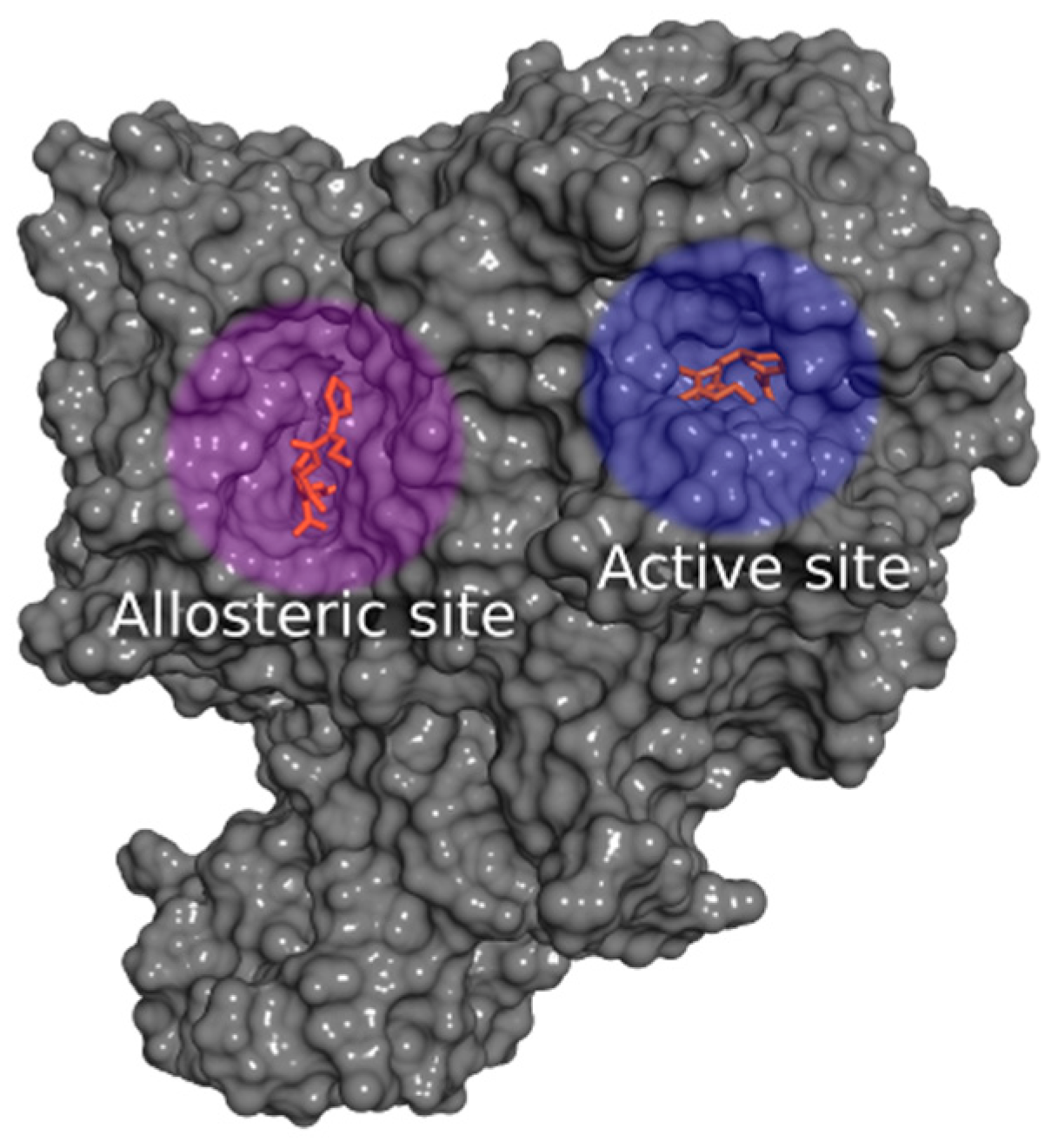
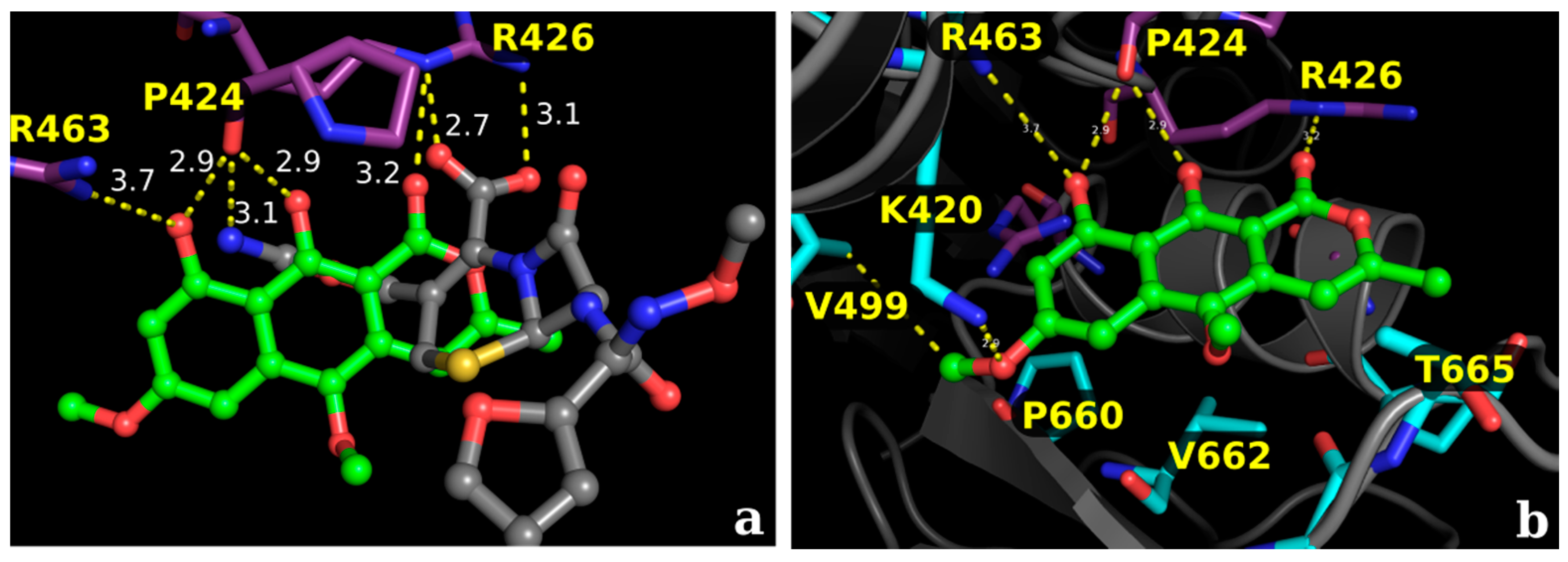
2.2.1. Docking Calculations
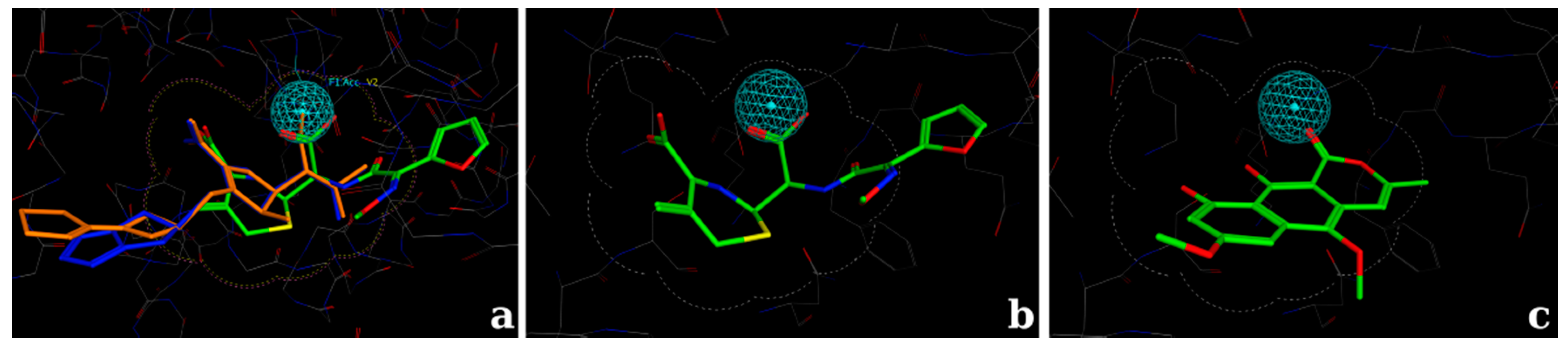
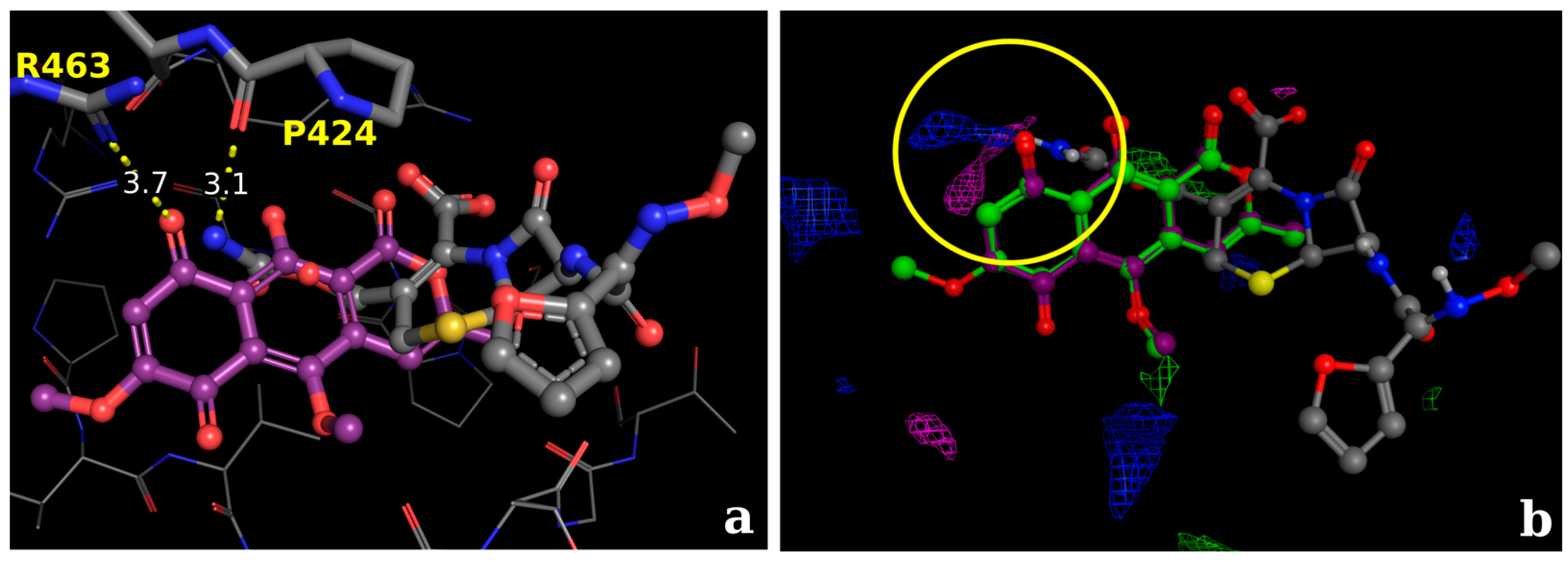
2.2.2. Docking Validation and Development of the Model
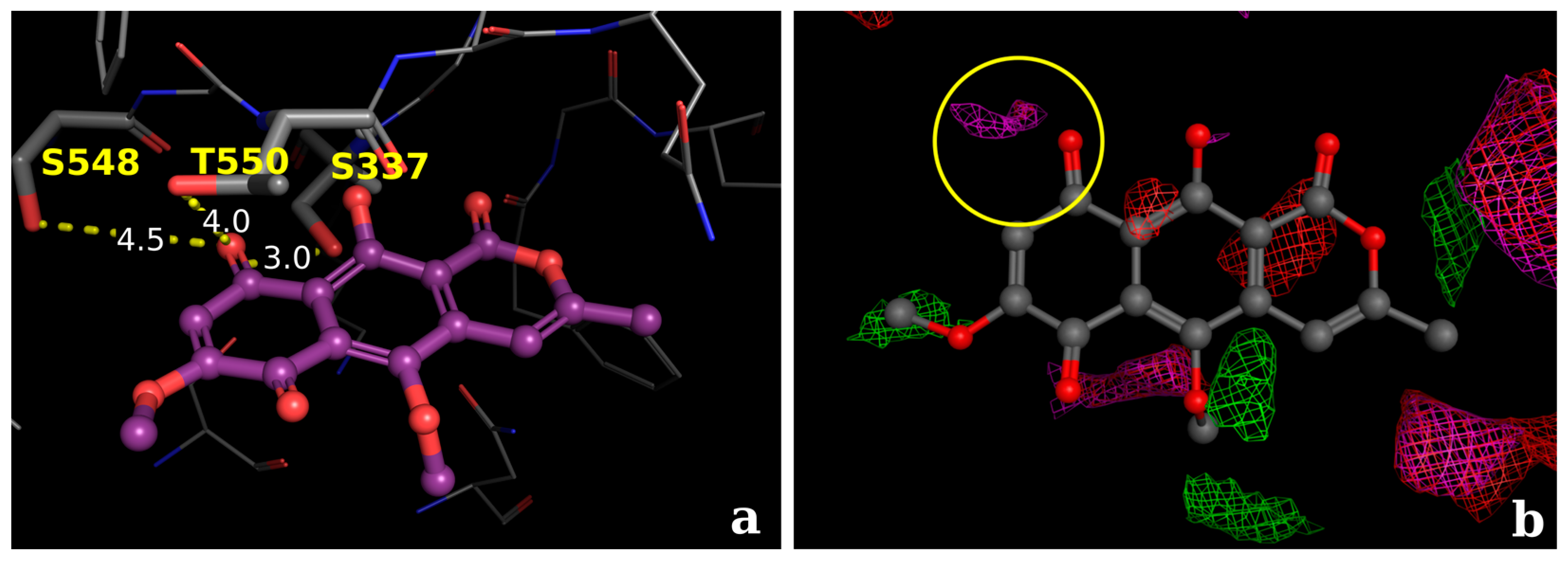
2.2.3. Docking Calculations for the Isocoumarin Paepalantine
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Chemicals
4.3. In Vitro Studies
4.3.1. Evaluation of Anti-H. pylori Activity
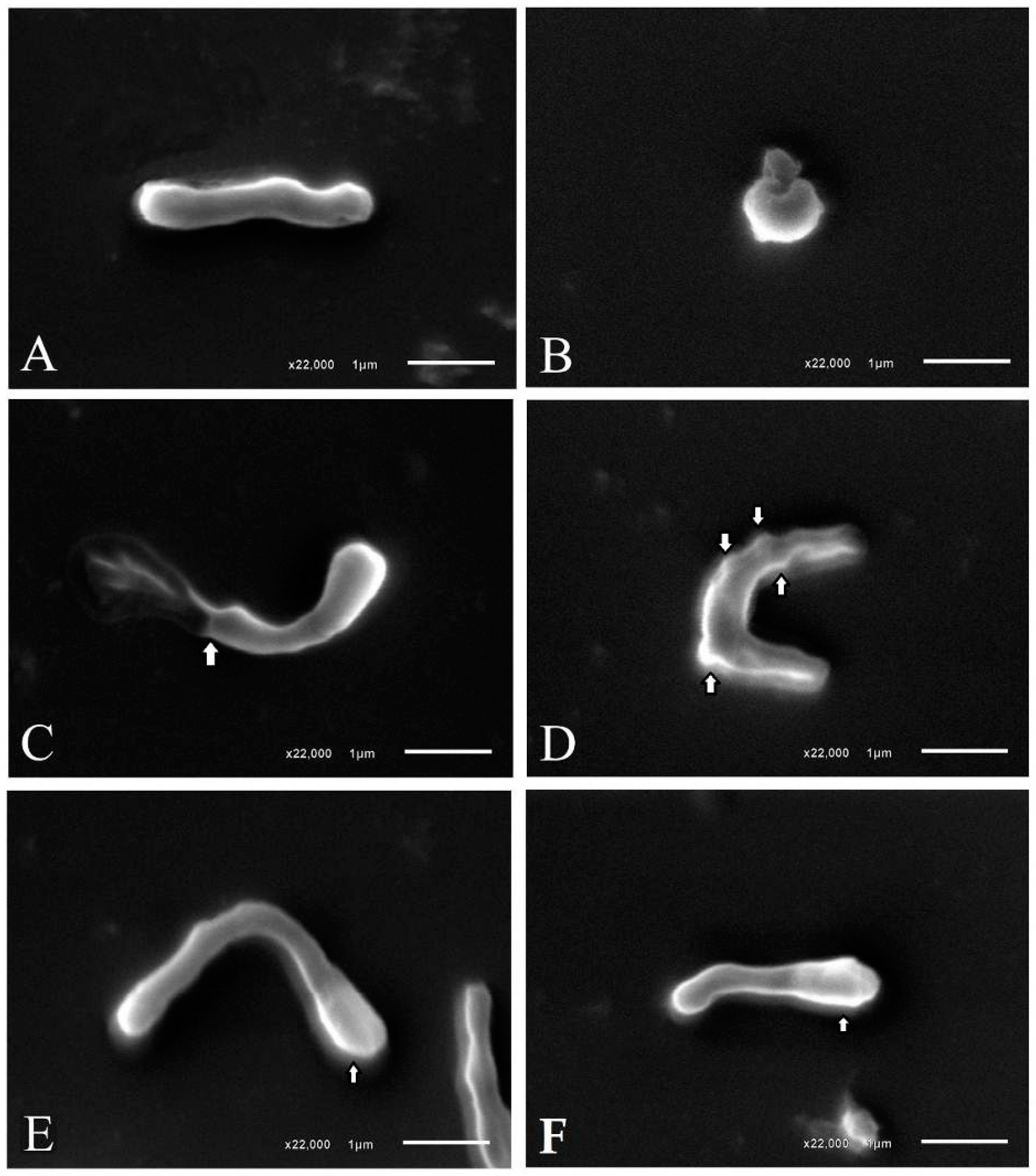
4.3.2. Morphological Analysis of H. pylori Bacterial Cells
4.4. Computational Methods
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ladeira, M.S.P.; Salvadori, D.M.F.; Rodrigues, M.A.M. Biopatologia do Helicobacter pylori. J. Bras. Patol. Med. Lab. 2003, 39, 335–342. [Google Scholar] [CrossRef]
- Kusters, J.G.; van Vliet, A.H.M.; Kuipers, E.J. Pathogenesis of Helicobacter pylori Infection. Clin. Microbiol. Rev. 2006, 19, 449–490. [Google Scholar] [CrossRef] [PubMed]
- Malaty, H.M. Epidemiology of Helicobacter pylori infection. Best Pract. Res. Clin. Gastroenterol. 2007, 21, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Cheng, N.; Dong, L.; Muramatsu, M.; Xiao, S.; Wang, M.W.; Zhu, D.X. Bactericidal and morphological effects of NE-2001, a novel synthetic agent directed against Helicobacter pylori. Antimicrob. Agents Chemother. 2005, 49, 3468–3473. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Alamuri, P.; Maier, R.J. The diverse antioxidant systems of Helicobacter pylori. Mol. Microbiol. 2006, 61, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Allen, L.A.H.; Beecher, B.R.; Lynch, J.T.; Rohner, O.V.; Wittine, L.M. Helicobacter pylori disrupts NADPH oxidase targeting in human neutrophils to induce extracellular superoxide release. J. Immunol. 2005, 174, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Antony, S.; Meitzler, J.L.; Doroshow, J.H. Molecular mechanisms underlying chronic inflammation-associated cancers. Cancer Lett. 2014, 345, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Eusebi, L.H.; Zagari, R.M.; Bazzoli, F. Epidemiology of Helicobacter pylori Infection. Helicobacter 2014, 19, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Miftahussurur, M.; Yamaoka, Y. Appropriate first-line regimens to combat Helicobacter pylori antibiotic resistance: An asian perspective. Molecules 2015, 20, 6068–6092. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Suzuki, H.; Suzuki, M.; Takahashi, M.; Hibi, T. Proton pump inhibitor-amoxicillin-clarithromycin versus proton pump inhibitor-amoxicillin-metronidazole as first-line Helicobacter pylori eradication therapy. J. Clin. Biochem. Nutr. 2012, 51, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.G.; Meshram, R.J.; Gond, D.S.; Gacche, R.N. Inhibition of growth of Helicobacter pylori and its urease by coumarin derivatives: Molecular docking analysis. J. Pharm. Res. 2013, 7, 705–711. [Google Scholar] [CrossRef]
- Bonacorsi, C.; Raddi, M.S.G.; Carlos, I.Z.; Sannomiya, M.; Vilegas, W. Anti-Helicobacter pylori activity and immunostimulatory effect of extracts from Byrsonima crassa Nied. (Malpighiaceae). BMC Complement. Altern. Med. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Ecclissato, C.; Marchioretto, M.A.M.; Mendonca, S.; Godoy, A.P.O.; Guersoni, R.A.; Deguer, M.; Piovesan, H.; Ferraz, J.G.P.; Pedrazzoli, J. Increased Primary Resistance to Recommended Antibiotics Negatively Affects Helicobacter pylori Eradication. Helicobacter 2002, 7, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Prasertpetmanee, S.; Mahachai, V.; Vilaichone, R.K. Improved efficacy of proton pump inhibitor-amoxicillin-clarithromycin triple therapy for Helicobacter pylori eradication in low clarithromycin resistance areas or for tailored therapy. Helicobacter 2013, 18, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Abu-Qatouseh, L.; Abu-Sini, M.; Mayyas, A.; Al-Hiari, Y.; Darwish, R.; Aburjai, T. Synthesis of New Nitrofluoroquinolone Derivatives with Novel Anti-Microbial Properties against Metronidazole Resistant H. pylori. Molecules 2017, 22, 71. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.; Anwar, F.; Naz, F.; Mehmood, T.; Saari, N. Anti-Helicobacter pylori and urease inhibition activities of some traditional medicinal plants. Molecules 2013, 18, 2135–2149. [Google Scholar] [CrossRef] [PubMed]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Davoine, F.; Lacy, P. Eosinophil cytokines, chemokines, and growth factors: Emerging roles in immunity. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Rob, J.A.; Tollefsen, S.; Helgeland, L. A Rapid and Highly Sensitive Chromogenic Microplate Assay for Quantification of Rat and Human Prothrombin. Anal. Biochem. 1997, 245, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Grover, J.; Jachak, S.M. Coumarins as privileged scaffold for anti-inflammatory drug development. RSC Adv. 2015, 5, 38892–38905. [Google Scholar] [CrossRef]
- Paya, M.; Goodwin, P.A.; De Las Heras, B.; Hoult, J.R.S. Superoxide scavenging activity in leukocytes and absence of cellular toxicity of a series of coumarins. Biochem. Pharmacol. 1994, 48, 445–451. [Google Scholar] [CrossRef]
- Kayser, O.; Kolodziej, H. Antibacterial activity of simple coumarins: Structural requirements for biological activity. Z. Naturforsch. C 1999, 54, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.V.C.; Kaplan, M.A.C. Tendências evolutivas de famílias produtoras de cumarinas em angiospermae. Quim. Nova 2002, 25, 533–538. [Google Scholar] [CrossRef]
- Hardy, C.D.; Cozzarelli, N.R. Alteration of Escherichia coli Topoisomerase IV to Novobiocin Resistance. Antimicrob. Agents Chemother. 2003, 47, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.G.; Cho, H.S.; Ryu, S.Y.; Cho, M.H.; Lee, J. Coumarins reduce biofilm formation and the virulence of Escherichia coli O157:H7. Phytomedicine 2014, 21, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.C.; Tsai, S.H.; Chen, C.S.; Chang, Y.C.; Lee, C.M.; Lai, Z.Y.; Lin, C.M. Structure–activity relationship of coumarin derivatives on xanthine oxidase-inhibiting and free radical-scavenging activities. Biochem. Pharmacol. 2008, 75, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Vilegas, W.; Roque, N.F.; Salatino, A.; Giesbrecht, A.M.; Davino, S. Isocoumarin from Paepalanthus bromelioides. Phytochemistry 1990, 29, 2299–2301. [Google Scholar] [CrossRef]
- Devienne, K.F.; Raddi, M.S.G. Screening for antimicrobial activity of natural products using a microplate photometer. Braz. J. Microbiol. 2002, 33, 166–168. [Google Scholar] [CrossRef]
- Ferrazzoli Devienne, K.; Gonçalves Raddi, M.S.; Gomes Coelho, R.; Vilegas, W. Structure-antimicrobial activity of some natural isocoumarins and their analogues. Phytomedicine 2005, 12, 378–381. [Google Scholar] [CrossRef]
- Kitagawa, R.R.; Raddi, M.S.G.; Khalil, N.M.; Vilegas, W.; da Fonseca, L.M. Effect of the isocoumarin paepalantine on the luminol and lucigenin amplified chemiluminescence of rat neutrophils. Biol. Pharm. Bull. 2003, 26, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Devienne, K.F.; Cálgaro-Helena, A.F.; Dorta, D.J.; Prado, I.M.R.; Raddi, M.S.G.; Vilegas, W.; Uyemura, S.A.; Santos, A.C.; Curti, C. Antioxidant activity of isocoumarins isolated from Paepalanthus bromelioides on mitochondria. Phytochemistry 2007, 68, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Di Stasi, L.C.; Camuesco, D.; Nieto, A.; Vilegas, W.; Zarzuelo, A.; Galvez, J. Intestinal anti-inflammatory activity of paepalantine, an isocoumarin isolated from the capitula of Paepalanthus bromelioides, in the trinitrobenzenesulphonic acid model of rat colitis. Planta Med. 2004, 70, 315–320. [Google Scholar] [PubMed]
- Varanda, E.A.; Devienne, K.F.; Raddi, M.S.G.; Furuya, E.M.; Vilegas, W. Mutagenicity of paepalantine dimer and glycoside derivatives from Paepalanthus bromelioides. Toxicol. In Vitro 2004, 18, 109–114. [Google Scholar] [CrossRef] [PubMed]
- DeLoney, C.R.; Schiller, N.L. Competition of various beta-lactam antibiotics for the major penicillin-binding proteins of Helicobacter pylori: Antibacterial activity and effects on bacterial morphology. Antimicrob. Agents Chemother. 1999, 43, 2702–2709. [Google Scholar] [PubMed]
- Berry, V.; Jennings, K.; Woodnutt, G. Bactericidal and Morphological Effects of Amoxicillin on Helicobacter-pylori. Antimicrob. Agents Chemother. 1995, 39, 1859–1861. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.; Meyer, E.F.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur. J. Biochem. 1977, 80, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Kirchmair, J.; Markt, P.; Distinto, S.; Schuster, D.; Spitzer, G.M.; Liedl, K.R.; Langer, T.; Wolber, G. The Protein Data Bank (PDB), Its Related Services and Software Tools as Key Components for In Silico Guided Drug Discovery. J. Med. Chem. 2008, 51, 7021–7040. [Google Scholar] [CrossRef] [PubMed]
- Mahasenan, K.V.; Molina, R.; Bouley, R.; Batuecas, M.T.; Fisher, J.F.; Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis. J. Am. Chem. Soc. 2017, 139, 2102–2110. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE) User Manual; Chemical Computing Group Inc.: Montreal, QC, Canada, 2013. [Google Scholar]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Plewczynski, D.; Łaźniewski, M.; Augustyniak, R.; Ginalski, K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J. Comput. Chem. 2011, 32, 742–755. [Google Scholar] [CrossRef] [PubMed]
- Selzer, P.M. Antiparasitic and Antibacterial Drug Discovery: From Molecular Targets to Drug Candidates; Wiley-Blackwell: Mörlenbach, Germany, 2009; Volume 1, p. 509. [Google Scholar]
- Vuorinen, A.; Schuster, D. Methods for generating and applying pharmacophore models as virtual screening filters and for bioactivity profiling. Methods 2015, 71, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Ayala, G.; Galván-Portillo, M.; Chihu, L.; Fierros, G.; Sánchez, A.; Carrillo, B.; Román, A.; López-Carrillo, L.; Silva-Sánchez, J.; Study Group. Resistance to Antibiotics and Characterization of Helicobacter pylori Strains Isolated from Antrum and Body from Adults in Mexico. Microb. Drug Resist. 2011, 17, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.Y.; Shiotani, A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat. Clin. Pract. Gastroenterol. Hepatol. 2008, 5, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Azab, A.; Nassar, A.; Azab, A.N. Anti-Inflammatory Activity of Natural Products. Molecules 2016, 21, 1321. [Google Scholar] [CrossRef] [PubMed]
- Vale, F.F. Overview of the phytomedicine approaches against Helicobacter pylori. World J. Gastroenterol. 2014, 20, 5594. [Google Scholar] [CrossRef] [PubMed]
- Vítor, J.M.B.; Vale, F.F. Alternative therapies for Helicobacter pylori: Probiotics and phytomedicine. FEMS Immunol. Med. Microbiol. 2011, 63, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Kostova, I.; Bhatia, S.; Grigorov, P.; Balkansky, S.; Parmar, V.S.; Prasad, A.K.; Saso, L. Coumarins as Antioxidants. Curr. Med. Chem. 2011, 18, 3929–3951. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ding, W.; Xu, Y.; Wu, D.; Li, S.; Chen, J.; Guo, B. New Insights into the Antibacterial Activity of Hydroxycoumarins against Ralstonia solanacearum. Molecules 2016, 21, 468. [Google Scholar] [CrossRef] [PubMed]
- Rauckman, B.S.; Tidwell, M.Y.; Johnson, J.V.; Roth, B. 2,4-Diamino-5-benzylpyrimidines and analogues as antibacterial agents. 10. 2,4-Diamino-5-(6-quinolylmethyl)- and -[(tetrahydro-6-quinolyl)methyl]pyrimidine derivatives. Further specificity studies. J. Med. Chem. 1989, 32, 1927–1935. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.; Mantoani, S.P.; de Almeida, J.R.; Pinsetta, F.R.; Semighini, E.P.; da Silva, V.B.; da Silva, C.H.T.P. Estratégias de Triagem Virtual no Planejamento de Fármacos. Rev. Virtual Quím. 2012, 4, 739–776. [Google Scholar]
- Kitagawa, R.R.; Bonacorsi, C.; da Fonseca, L.M.; Vilegas, W.; Raddi, M.S.G. Anti-Helicobacter pylori activity and oxidative burst inhibition by the naphthoquinone 5-methoxy-3,4-dehydroxanthomegnin from Paepalanthus latipes. Rev. Bras. Farmacogn. 2012, 22, 53–59. [Google Scholar] [CrossRef]
- Damasceno, J.P.L.; dos Giuberti, C.S.; de Gonçalves, R.C.R.; Kitagawa, R.R. Preformulation study and influence of DMSO and propylene glycol on the antioxidant action of isocoumarin paepalantine isolated from Paepalanthus bromelioides. Rev. Bras. Farmacogn. 2015, 25, 395–400. [Google Scholar] [CrossRef]
- Chakraborti, S.; Bhattacharya, S.; Chowdhury, R.; Chakrabarti, P. The molecular basis of inactivation of metronidazole-resistant Helicobacter pylori using polyethyleneimine functionalized zinc oxide nanoparticles. PLoS ONE 2013, 8, e70776. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.L.; Walters, K.S.; Drake, D.R.; Blanchette, D.R.; Dawson, D.V.; Brogden, K.A.; Wertz, P.W. Sphingoid Bases Are Taken Up by Escherichia coli and Staphylococcus aureus and Induce Ultrastructural Damage. Skin Pharmacol. Physiol. 2013, 26, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Claverie-Martin, F.; Diaz-Torres, M.R.; Geoghegan, M.J. Chemical composition and ultrastructure of wild-type and white mutantAspergillus nidulans conidial walls. Curr. Microbiol. 1988, 16, 281–287. [Google Scholar] [CrossRef]
- ChemAxon Marvin Sketch. Available online: https://www.chemaxon.com/products/marvin/marvinsketch/ (accessed on 21 December2016).
- Vainio, M.J.; Johnson, M.S. Generating conformer ensembles using a multiobjective genetic algorithm. J. Chem. Inf. Model. 2007, 47, 2462–2474. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- OMEGA 2.5.1.4: OpenEye Scientific Software, Santa Fe, NM. Available online: http://www.eyesopen.com (accessed on 9 January 2017).
- Wilke, M.S.; Lovering, A.L.; Strynadka, N.C. β-Lactam antibiotic resistance: A current structural perspective. Curr. Opin. Microbiol. 2005, 8, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.M.; Anbarasu, A.; Ramaiah, S. Molecular docking and molecular dynamics studies on β-lactamases and penicillin binding proteins. Mol. Biosyst. 2014, 10, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Im, H.; Lee, K.Y.; Chen, J.; Kang, H.J.; Yoon, H.J.; Min, K.H.; Lee, K.R.; Park, H.J.; Lee, B.J. Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS). Eur. J. Med. Chem. 2016, 108, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Fock, K.M.; Graham, D.Y.; Malfertheiner, P. Helicobacter pylori research: Historical insights and future directions. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 495–500. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Compound samples can be available from the authors. |


| PDB Code | Target | Inhibitor | Class | Reference |
|---|---|---|---|---|
| 1QMF | PBP2x | Cefuroxime | Cephalosporin | doi:10.1006/jmbi.2000.3740 |
| 2ZC4 | PBP2x | Tebipenem | Carbapenem | doi:10.1128/AAC.01456-07 |
| 2ZC3 | PBP2x | Biapenem | Carbapenem | doi:10.1128/AAC.01456-07 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damasceno, J.P.L.; Rodrigues, R.P.; Gonçalves, R.D.C.R.; Kitagawa, R.R. Anti-Helicobacter pylori Activity of Isocoumarin Paepalantine: Morphological and Molecular Docking Analysis. Molecules 2017, 22, 786. https://doi.org/10.3390/molecules22050786
Damasceno JPL, Rodrigues RP, Gonçalves RDCR, Kitagawa RR. Anti-Helicobacter pylori Activity of Isocoumarin Paepalantine: Morphological and Molecular Docking Analysis. Molecules. 2017; 22(5):786. https://doi.org/10.3390/molecules22050786
Chicago/Turabian StyleDamasceno, João Paulo L., Ricardo P. Rodrigues, Rita De Cássia R. Gonçalves, and Rodrigo R. Kitagawa. 2017. "Anti-Helicobacter pylori Activity of Isocoumarin Paepalantine: Morphological and Molecular Docking Analysis" Molecules 22, no. 5: 786. https://doi.org/10.3390/molecules22050786