Antifungal Activity of Ramulus cinnamomi Explored by 1H-NMR Based Metabolomics Approach
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure of Compounds 1–2
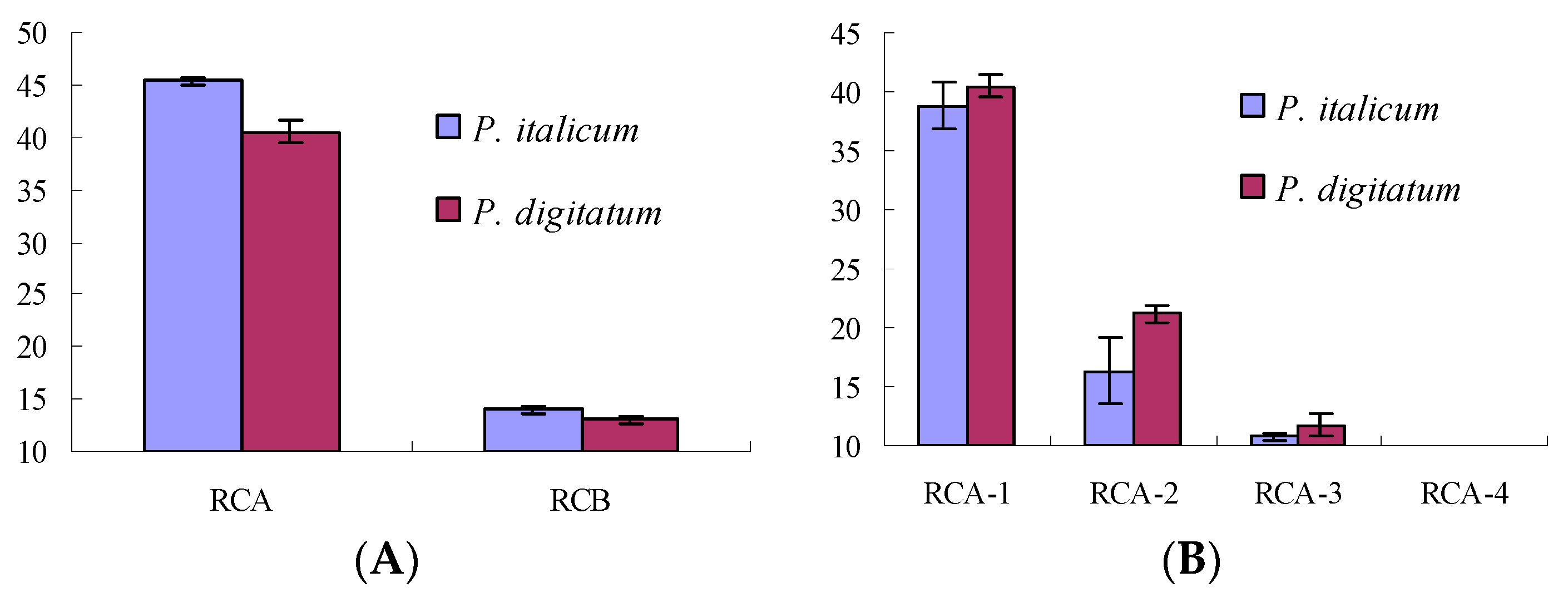
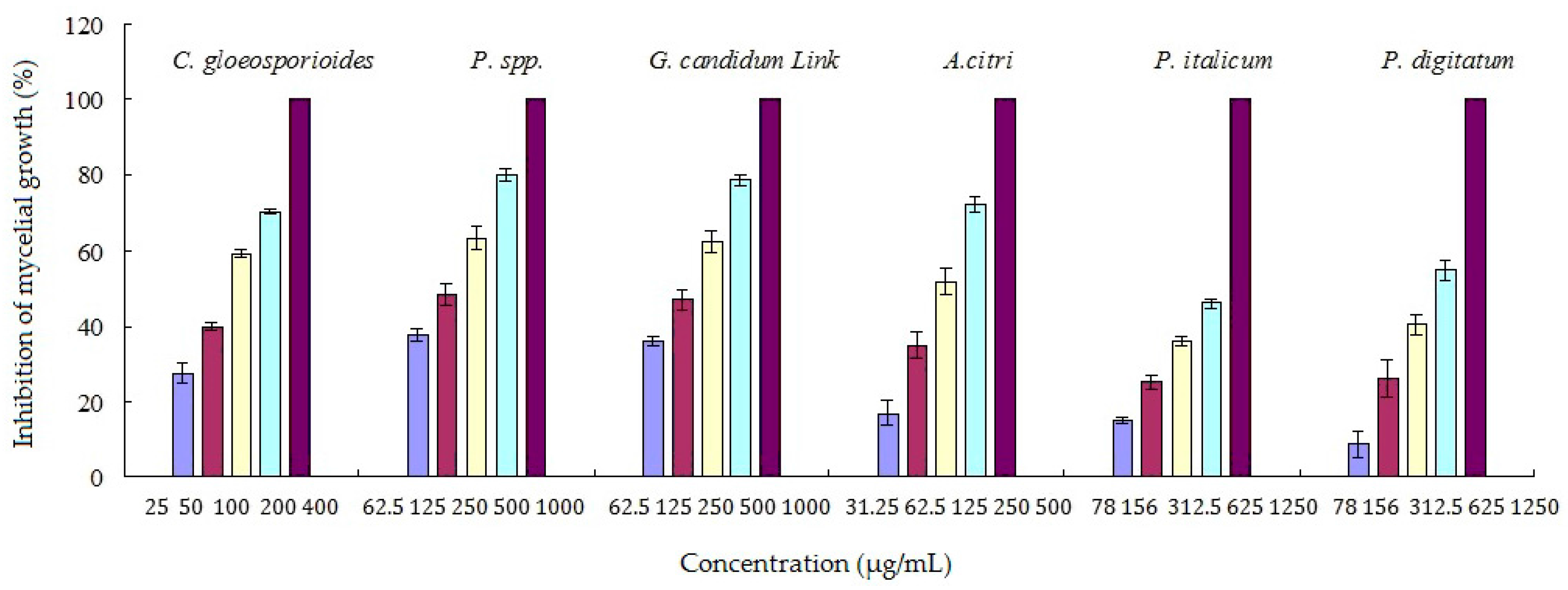
2.2. Antifungal Activities of RC Extracts and Fractions
2.3. Antifungal Activity of Cinnamaldehyde
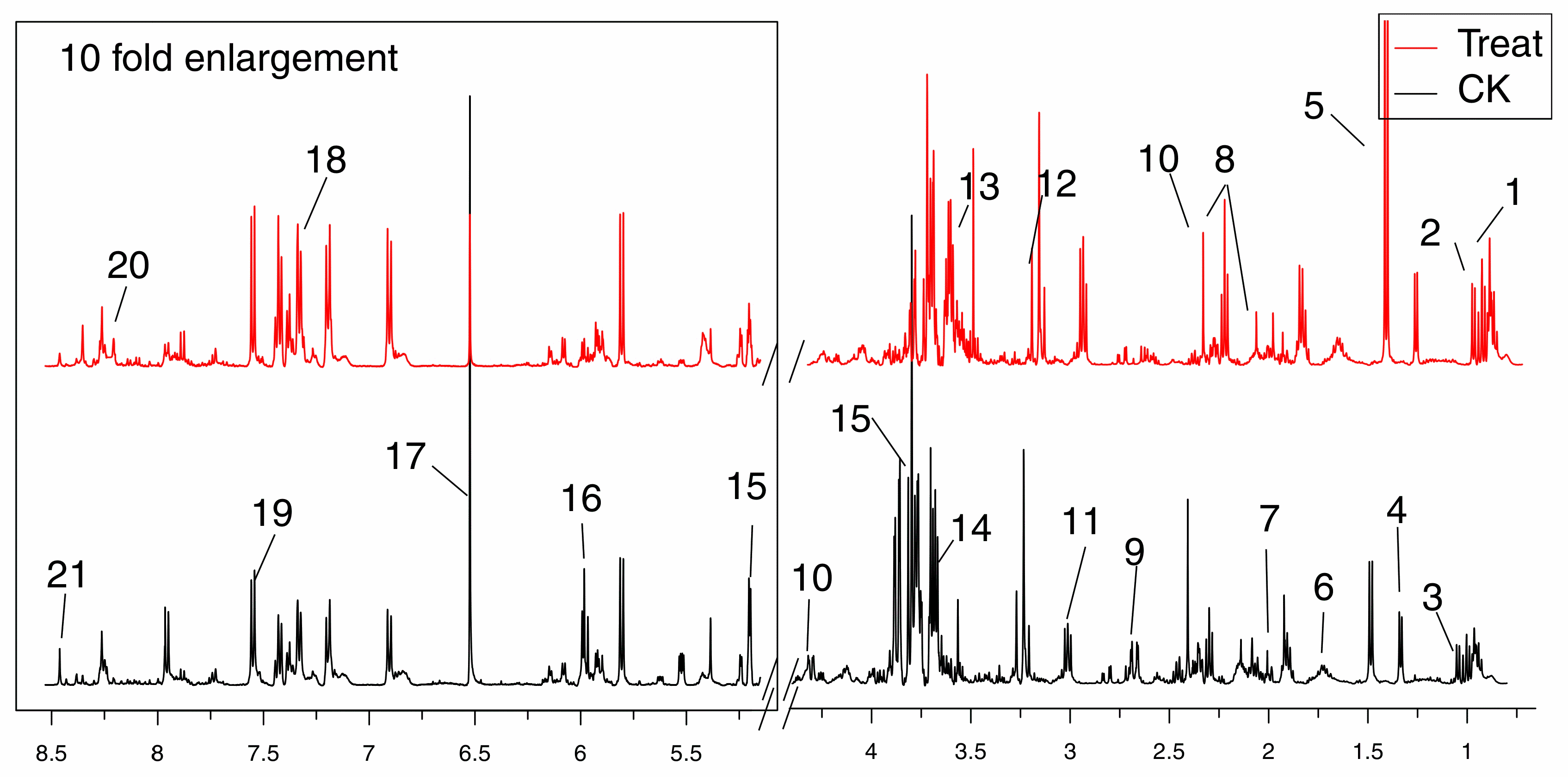
2.4. Metabolites Identified in 1H-NMR Spectra
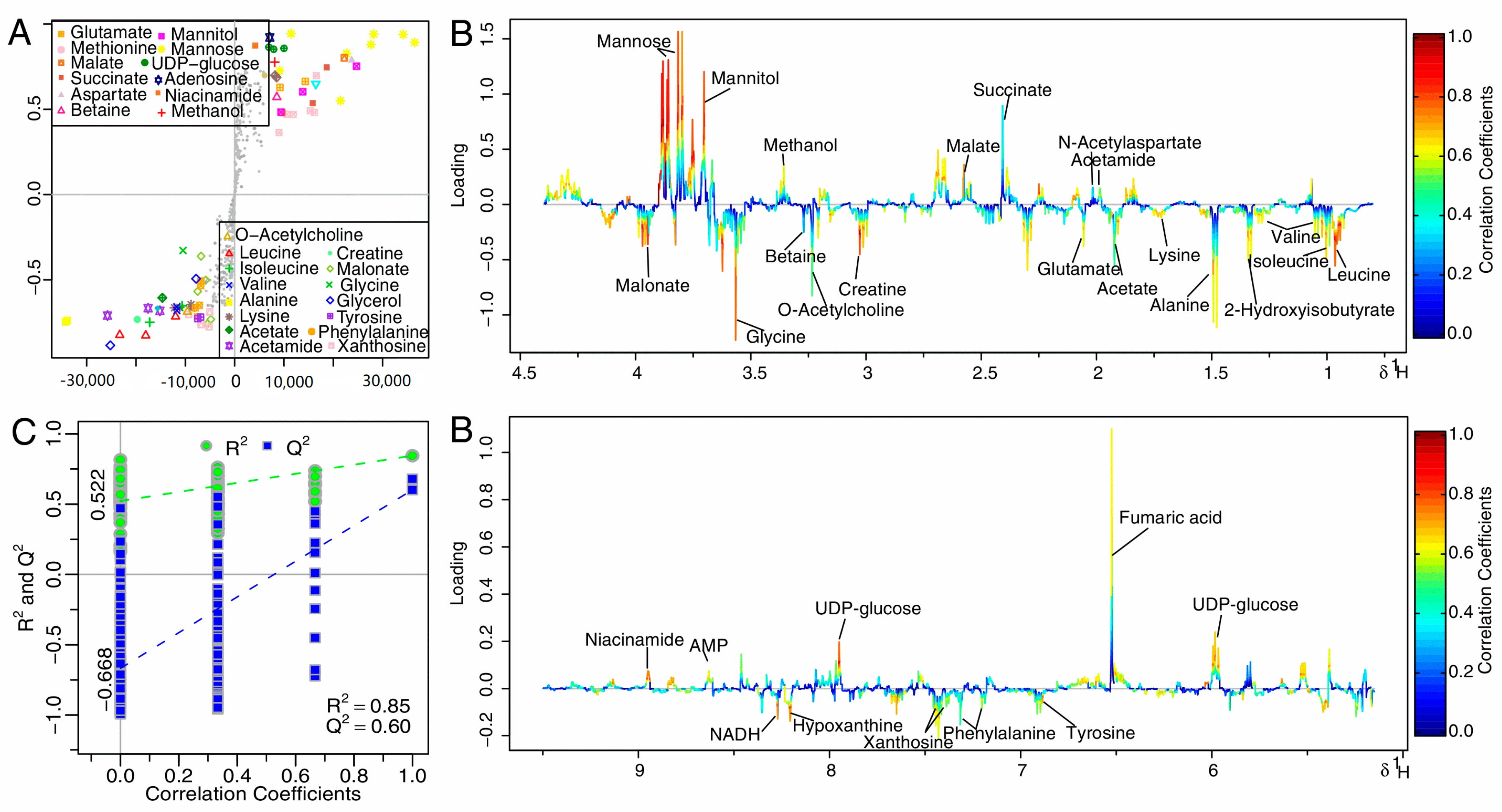
2.5. Multivariate Analysis of 1H-NMR Spectral Data
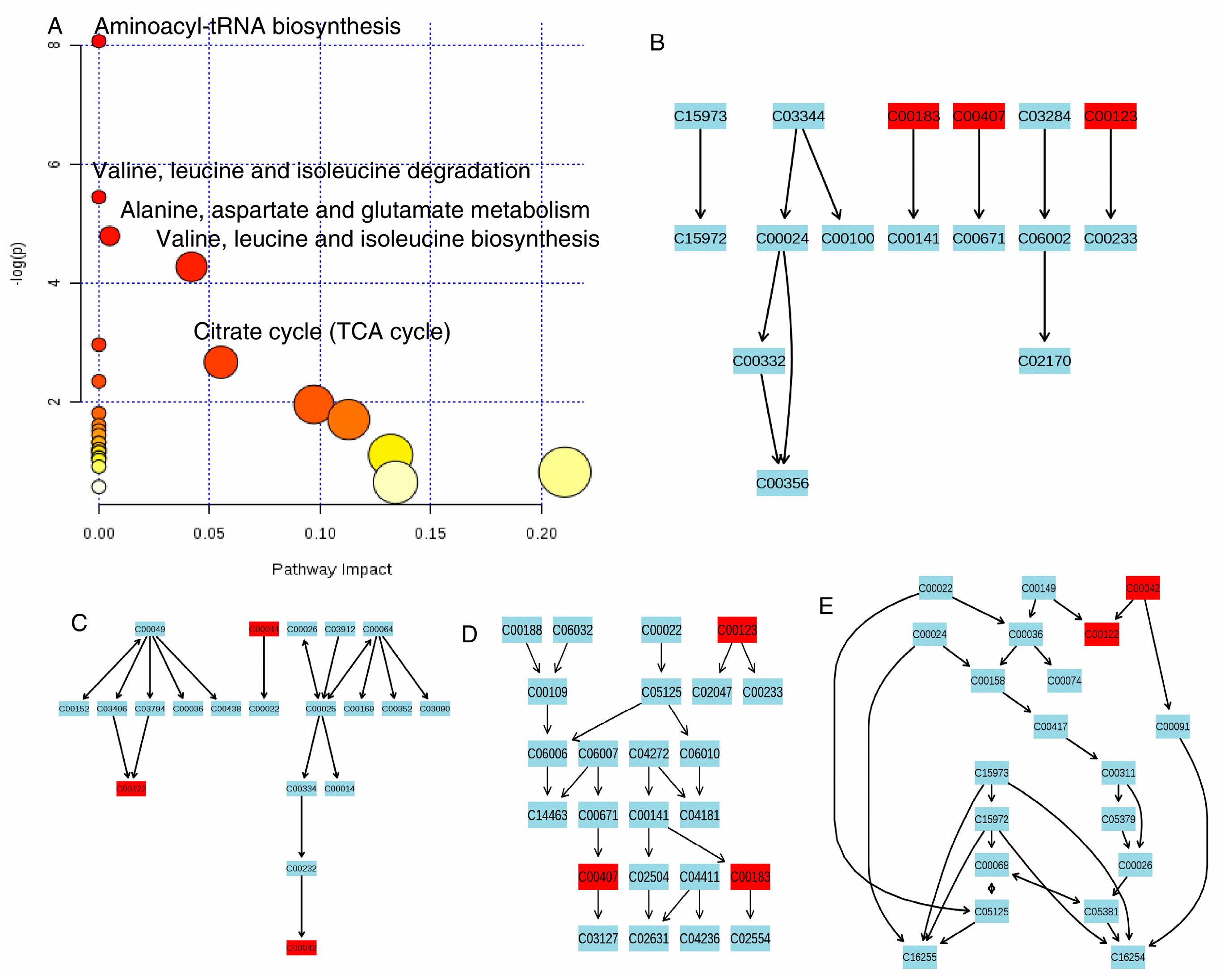
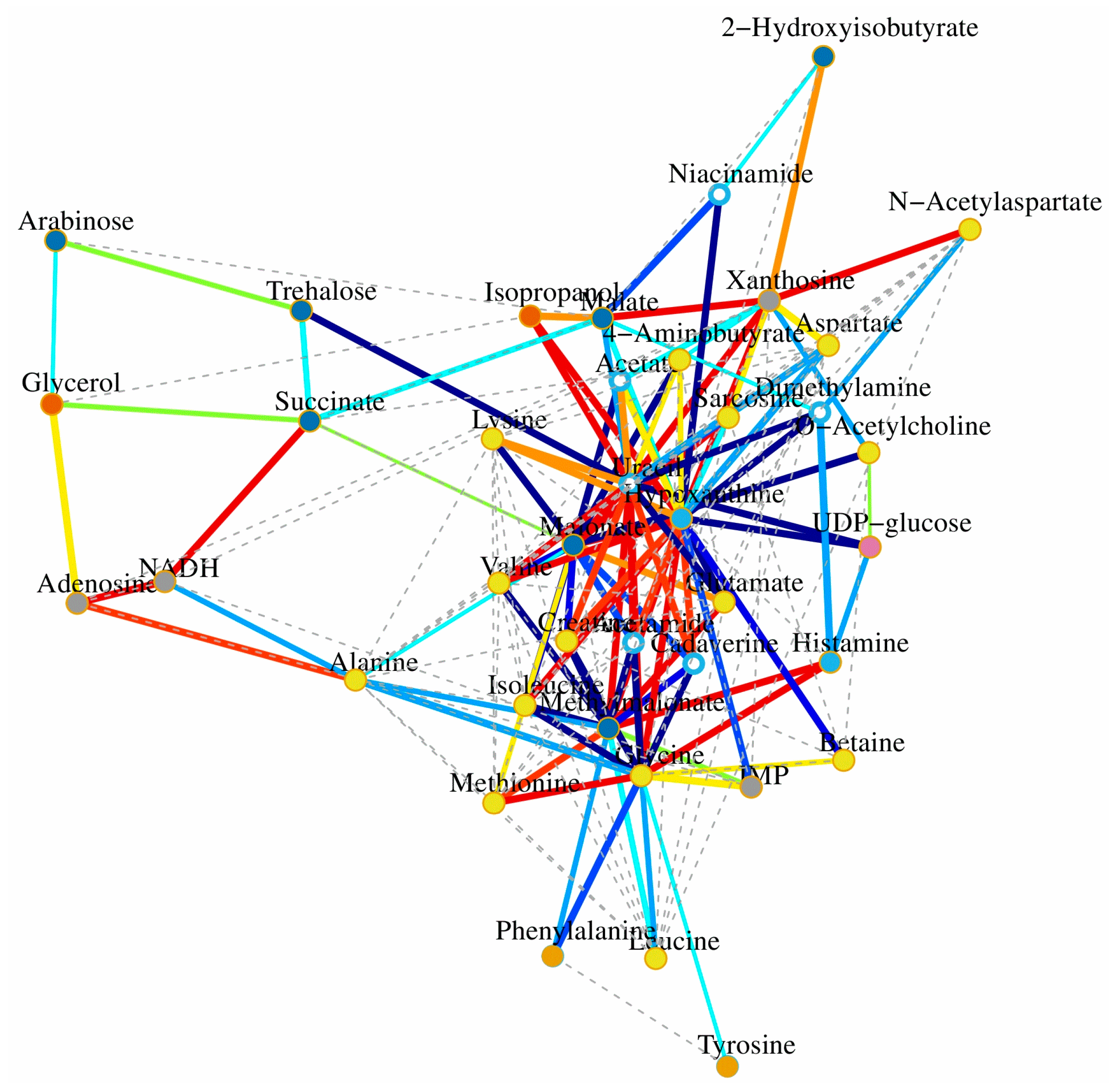
2.6. Metabolomics Analysis
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Extraction and Isolation
3.3. Antifungal Activity Tested by the Oxford Cup Method
3.4. Antifungal Activity Tested by the Mycelial Growth Method
3.5. 1H-NMR Based Metabolomics Analysis
3.6. Spectral Pre-Processing and Data Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NMR | Nuclear magnetic resonance |
| 1H-NMR | 1H nuclear magnetic resonance |
| 13C-NMR | 13C nuclear magnetic resonance |
| NOESY | Nuclear overhauser enhancement spectroscopy |
| RC | Ramulus cinnamomi |
| RCA | Extraction of Ramulus cinnamomi by ethyl acetate |
| RCB | Extraction Ramulus cinnamomi by n-buthanol |
| OSC-PLS-DA | Orthogonal signal correction partial least-squares discriminant analysis |
| DIZs | Diameters of inhibition zones |
| HMDB | Human Metabolome Database |
| MMCD | Madison-Qingdao Metabolomics Consortium Database |
| STOCSY | Statistical total correlation spectroscopy |
| sPLS | Sparse-partial least-squares |
| BCAAs | Branched-chain amino acids |
| AAAs | Aromatic amino acids |
| NAA | N-acetylaspartate |
| GSH | glutathione |
| ROS | Reactive oxygen species |
| HPLC | High Performance Liquid Chromatography |
| PDA | Potato Dextrose Agar |
| PDB | Potato Dextrose Broth |
| IMG | Inhibition of mycelial growth |
| CFU | Colony-Forming Units |
| PBS | Phosphate buffered solution |
| PQN | Probability quotient normalization |
References
- Strano, M.C.; Di Silvestro, S.; Coniglione, M.; Lio, S.; Magnano, R. Decay control of cold stored Citrus clementina Hort. ex Tan. fruit by pre-and postharvest application of potassium phosphite. Adv. Hortic. Sci. 2015, 2–3, 97–102. [Google Scholar]
- Zhou, M.J.; Wan, C.P.; Chen, J.Y. Screening and antifungal mechanism of Chinese herb extracts against green mold of citrus. Mod. Food Sci. Technol. 2014, 3, 144–149. [Google Scholar]
- Wan, C.P.; Zhou, M.J.; Liu, Y.; Chen, J.Y. Antifungal activity of Ramulus cinnamomi extracts against two key citrus postharvest pathogens. Acta Agric. Univ. Jiangxiensis 2014, 2, 319–325. [Google Scholar]
- Chen, Y.H.; Peng, X.; Chen, C.Y.; Wan, C.P.; Chen, J.Y. Induction of blue mold resistance and defense system in Xinyu tangerine Ramulus cinnamomi main active ingredients. J. Plant Prot. 2016, 3, 467–474. [Google Scholar]
- Liang, M.T.; Yang, C.H.; Li, S.T.; Yang, C.S.; Chang, H.W.; Liu, C.S.; Chuang, L.Y. Antibacterial and antioxidant properties of Ramulus Cinnamomi using supercritical CO2 extraction. Eur. Food. Res. Technol. 2008, 5, 1387–1396. [Google Scholar] [CrossRef]
- Jung, J.; Lee, J.H.; Bae, K.H.; Jeong, C.S. Anti-gastric actions of eugenol and cinnamic acid isolated from Cinnamomi Ramulus. Yakugaku Zasshi 2011, 7, 1103–1110. [Google Scholar] [CrossRef]
- Wang, X.; Jia, W.; Zhao, A.; Wang, X. Anti-influenza agents from plants and traditional Chinese medicine. Phytother. Res. 2006, 5, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Shin, H.M. Cinnamomi ramulus ethanol extract exerts vasorelaxation through inhibition of influx and release in rat aorta. Evid.-Based Complement. Altern. Med. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Han, J.; Chen, C.; Yao, L.; Chen, J.; Yuan, T. Monosubstituted benzene derivatives from fruits of Ficus hirta and their antifungal activity against phytopathogen Penicillium italicum. J. Agric. Food Chem. 2016, 28, 5621–5624. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wan, C.; Peng, X.; Chen, Y.; Chen, M.; Chen, J. Optimization of antifungal extracts from Ficus hirta fruits using response surface methodology and antifungal activity tests. Molecules 2015, 11, 19647–19659. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Peng, X.; Zeng, R.; Chen, M.; Wan, C.; Chen, J. Ficus hirta fruits extract incorporated into an alginate-based edible coating for Nanfeng mandarin preservation. Sci. Hortic. 2016, 202, 41–48. [Google Scholar] [CrossRef]
- Chen, C.Y.; Zheng, J.P.; Wan, C.P.; Chen, M.; Chen, J.Y. Effect of carboxymethyl cellulose coating enriched with clove oil on postharvest quality of ‘Xinyu’ mandarin oranges. Fruits 2016, 5, 319–327. [Google Scholar]
- Xu, Q.; Zhu, J.; Wang, Z.; Xiao, W. α-Glucosidase inhibitors from Cinnamomi Cortex. Nat. Prod. Res. Dev. 2012, 9, 1246–1249. [Google Scholar]
- Alvaro, M.; Maria, J.; Javier, F.; Jimenez-Barbero, J. Structure and function of prokaryotic UDP-Glucose pyrophosphorylase, a drug target candidate. Curr. Med. Chem. 2015, 14, 1687–1697. [Google Scholar]
- Wu, C.W.; Wu, X.J.; Wen, C.; Peng, B.; Peng, X.X.; Chen, X.; Li, H. Fructose promotes growth and antifungal activity of Penicillium citrinum. Protein Cell 2016, 7, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Cramer, F.; Englisch, U.; Freist, W.; Sternbach, H. Aminoacylation of tRNAs as critical step of protein biosynthesis. Biochimie 1991, 7, 1027–1035. [Google Scholar] [CrossRef]
- Lu, L.F.; Wang, J.X.; Zhu, R.Y.; Lu, H.; Zheng, X.; Yu, T. Transcript profiling analysis of Rhodosporidium paludigenum-mediated signalling pathways and defense responses in mandarin orange. Food Chem. 2015, 172, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.R.; Liu, H.L.; Wu, J.F.; Zhang, X.; Liu, M.; Wang, Y. Metabonomic alterations in hippocampus, temporal and prefrontal cortex with age in rats. Neurochem. Int. 2009, 8, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Thangamani, S.; Eldesouky, H.E.; Mohammad, H.; Pascuzzi, P.E.; Avramova, L.; Hazbun, T.R.; Seleem, M.N. Ebselen exerts antifungal activity by regulating glutathione (GSH) and reactive oxygen species (ROS) production in fungal cells. BBA-Gen. Subj. 2017, 1861, 3002–3010. [Google Scholar] [CrossRef] [PubMed]
- Icksoo, L. Betaine is a positive regulator of mitochondrial respiration. Biochem. Biophys. Res. Commun. 2015, 2, 621–625. [Google Scholar]
- Goudela, S.; Reichard, U.; Amillis, S.; Diallinas, G. Characterization and kinetics of the major purine transporters in Aspergillus fumigatus. Fungal Genet. Biol. 2008, 4, 459–472. [Google Scholar] [CrossRef] [PubMed]
- De Koning, W.; van Dam, K. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal. Biochem. 1992, 204, 118–123. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, C.; Li, P.; Chen, C.; Peng, X.; Li, M.; Chen, M.; Wang, J.; Chen, J. Antifungal Activity of Ramulus cinnamomi Explored by 1H-NMR Based Metabolomics Approach. Molecules 2017, 22, 2237. https://doi.org/10.3390/molecules22122237
Wan C, Li P, Chen C, Peng X, Li M, Chen M, Wang J, Chen J. Antifungal Activity of Ramulus cinnamomi Explored by 1H-NMR Based Metabolomics Approach. Molecules. 2017; 22(12):2237. https://doi.org/10.3390/molecules22122237
Chicago/Turabian StyleWan, Chunpeng, Pei Li, Chuying Chen, Xuan Peng, Mingxi Li, Ming Chen, Junsong Wang, and Jinyin Chen. 2017. "Antifungal Activity of Ramulus cinnamomi Explored by 1H-NMR Based Metabolomics Approach" Molecules 22, no. 12: 2237. https://doi.org/10.3390/molecules22122237





