An Interface for Biomedical Big Data Processing on the Tianhe-2 Supercomputer
Abstract
:1. Introduction
- We analyze the need for and possibility of convenient big data support on modern supercomputers
- We propose Orion, which is an easy-to-use interface for users who wants to run Hadoop/Spark applications on the Tianhe-2 supercomputer
- We test Orion on Tianhe-2 against an in-house Hadoop cluster using the genomics big data example and testify the utility of Orion
2. Results and Discussion
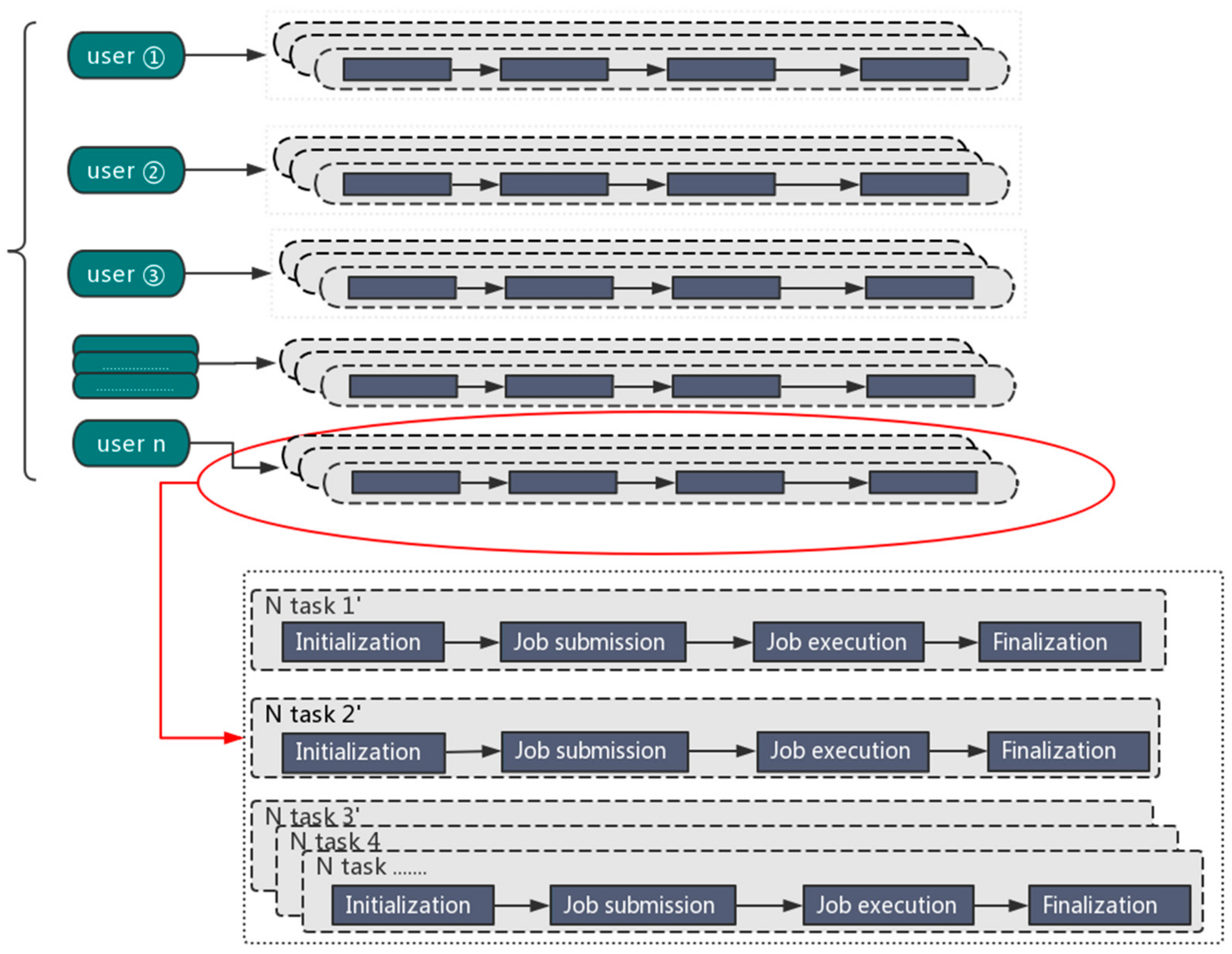
2.1. Automated Deployment of Big Data Applications
2.2. Testing Orion: Genomics Big Data as a Case Study
2.3. Portability of the Interface
3. Materials and Methods
3.1. Features of Orion
- It supports Hadoop and Spark configuration automatically; the user only needs to specify the framework type and the number of nodes.
- We use the original scheduling system of Tianhe-2 and do not use YARN for node management.
- We use the H2FS filesystem of Tianhe-2 instead of HDFS.
- We not only configure the framework work environment, but also enable related environments like NoSQL databases support (Redis, Mongo DB), OpenMP, and MPI.
- The Tianhe-2 monitoring subsystem can track the status of the temporarily created big data analytics cluster dynamically.
- The temporary data analytics cluster can be created and recycled any time the users wish, so that users will not be charged if their applications are not actually running.
3.2. Demonstration of Orion Usage (Tianhe-2 in the NSCC Guangzhou as an Example)
- Log in to your user account on Tianhe-2 and you will be given a command line terminal. Then, you can see the following:
- The installation directory
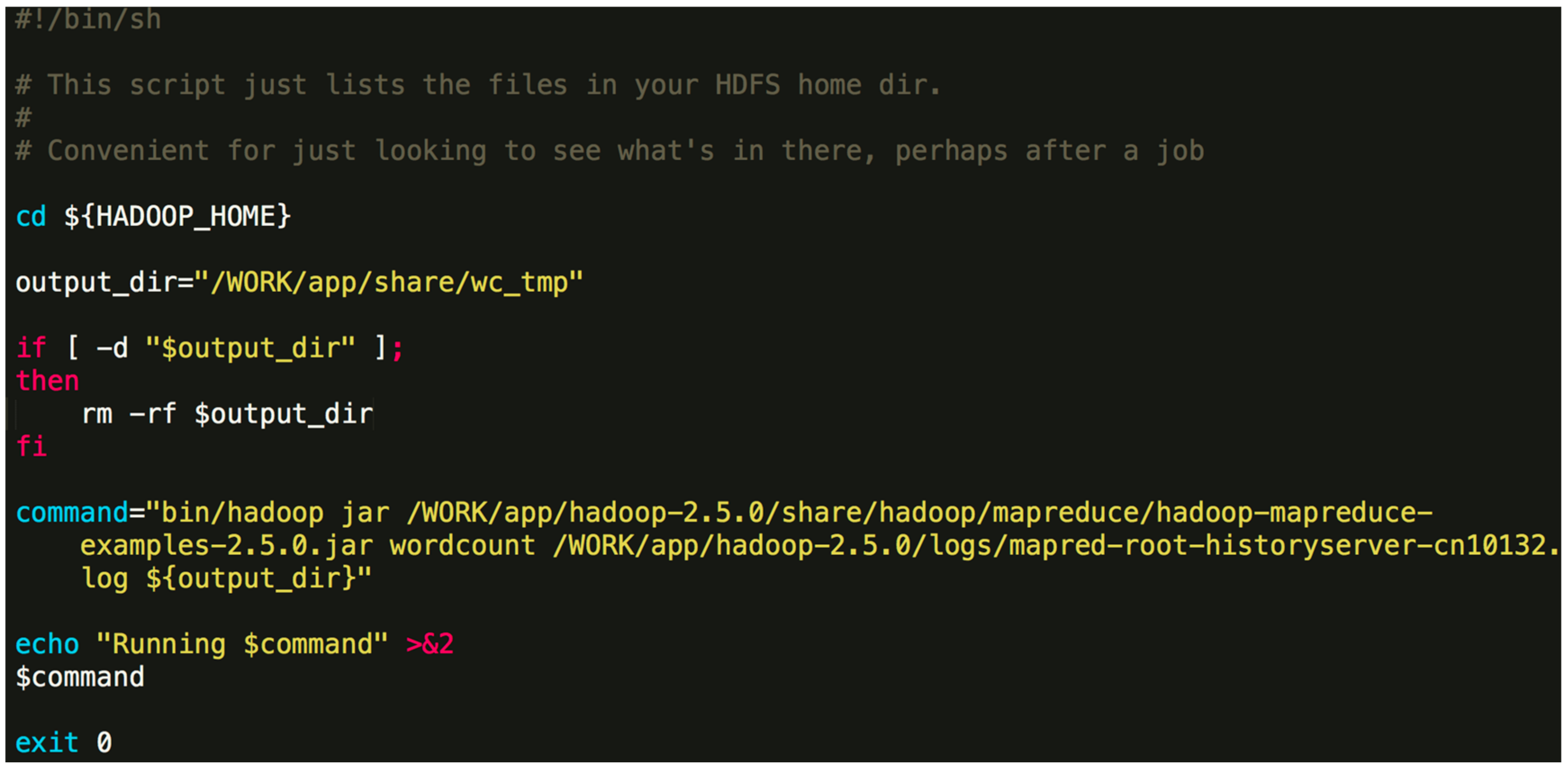
/WORK/app/share/Orion - The initial situation in this directory is as the Figure 3.Orion_start.sh is used for cluster configuration and start. Orion_clean.sh is the task run after the completion of the calculation or the release of resources upon an error, and cleans up the directory. Script directory is used to store task scripts.
- Switch to the Orion installation directory and execute a command like:
./Orion_start.sh -t hadoop -p work -N 6 -s ‘pwd’/script/wordcount.sh Here, the “-t” option can either be “hadoop” or “spark”, which specifies the big data framework you are using; the “-p” option specifies the resource pool on Tianhe-2 you want to use; the “-N” option specifies the number of compute nodes to use; the “-s” option specifies the job Shell script to be executed. The first three options are obligatory. The last one is optional. An example job script “wordcount.sh” is given in Figure 4.The above command will invoke the initialization process and wait for the resources to be allocated. If the required resources cannot be allocated after a long time, Orion will prompt the user to select a different resource pool or to use less nodes.$ {HADOOP_HOME} -points to the directory where Hadoop is installed. In general, the contents of the command can be modified. You can also specify a different output directory. - Once the task is submitted, it will be automatically queued to wait for resources. If the designated resource requirements cannot be fulfilled, Orion will give a message and you will need to switch to a partition with more idle resources or reduce the number of nodes.
- If the application for resource is successful, Orion will continue to configure and launch a Hadoop/Spark cluster using the allocated nodes.
- Orion executes the designated script and waits for the execution to finish. You can continue to use the configured cluster to run jobs interactively.
- Use a command like the following to terminate the cluster and release occupied resources:
./Orion_clean.sh -j 1362049 Here the “-j” option tells Orion to clean up the corresponding job, and the job ID is given in Step 2.
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tolle, K.M.; Tansley, D.S.W.; Hey, A.J.G. The fourth paradigm: Data-intensive scientific discovery. Proc. IEEE 2011, 99, 1334–1337. [Google Scholar] [CrossRef]
- Marx, V. Biology: The big challenges of big data. Nature 2013, 498, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Zikopoulos, P.; Eaton, C. Understanding Big Data: Analytics for Enterprise Class Hadoop and Streaming Data; McGraw-Hill Osborne Media: Columbus, OH, USA, 1989. [Google Scholar]
- Shvachko, K.; Kuang, H.; Radia, S.; Chansler, R. The Hadoop distributed file system. In Proceedings of the 2010 IEEE 26th Symposium on Mass Storage Systems and Technologies (MSST), Incline Village, NV, USA, 3–7 May 2010; pp. 1–10. [Google Scholar]
- Zou, Q.; Li, X.; Jiang, W.; Lin, Z.; Li, G.; Chen, K. Survey of MapReduce frame operation in bioinformatics. Brief. Bioinform. 2017, 15, 637–647. [Google Scholar] [CrossRef]
- Zou, Q.; Hu, Q.; Guo, M.; Wang, G. HAlign : Fast multiple similar DNA/RNA sequence alignment based on the centre star strategy. Bioinformatics 2017, 31, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, R. Hadoop MapReduce framework to implement molecular docking of large-scale virtual screening. In Proceedings of the Services Computing Conference (APSCC), 2012 IEEE Asia-Pacific, Guilin, China, 6–8 December 2012; pp. 350–353. [Google Scholar]
- Zhang, Y.; Zhang, R.; Chen, Q.; Gao, X.; Hu, R.; Zhang, Y.; Liu, G. A hadoop-based massive molecular data storage solution for virtual screening. In Proceedings of the 2012 Seventh China Grid Annual Conference, Beijing, China, 20–23 September 2012. [Google Scholar]
- Niu, J.; Bai, S.; Khosravi, E.; Park, S. A Hadoop approach to advanced sampling algorithms in molecular dynamics simulation on cloud computing. In Proceedings of the 2013 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shanghai, China, 18–21 December 2013; pp. 452–455. [Google Scholar]
- Li, J.; Li, D.; Zhang, Y. Efficient distributed data clustering on spark. In Proceedings of the 2015 IEEE International Conference on Cluster Computing (CLUSTER), Chicago, IL, USA, 8–11 September 2015; pp. 504–505. [Google Scholar]
- Yang, X.; Liao, X.; Lu, K.; Hu, Q.; Song, J.; Su, J. The TianHe-1A supercomputer: Its hardware and software. Comput. Sci. 2011, 26, 344–351. [Google Scholar] [CrossRef]
- Fascio, V.; Wüthrich, R.; Bleuler, H. Spark assisted chemical engraving in the light of electrochemistry. Electrochim. Acta 2004, 49, 3997–4003. [Google Scholar] [CrossRef]
- Zaharia, M.; Chowdhury, M.; Franklin, M.J.; Shenker, S.; Stoica, I. Spark: Cluster computing with working sets. In Proceedings of the 2nd USENIX Conference on Hot Topics in Cloud Computing, Boston, MA, USA, 22–25 June 2010; p. 10. [Google Scholar]
- Liao, X.; Xiao, L.; Yang, C.; Lu, Y. MilkyWay-2 supercomputer: System and application. Front. Comput. Sci. 2014, 8, 345–356. [Google Scholar] [CrossRef]
- Cheptsov, A. HPC in big data age: An evaluation report for java-based data-intensive applications implemented with Hadoop and OpenMPI. In Proceedings of the 21st European MPI Users’ Group Meeting, Kyoto, Japan, 9–12 September 2014; p. 175. [Google Scholar]
- Islam, N.S.; Shankar, D.; Lu, X.; Panda, D.K. Accelerating I/O performance of big data analytics on HPC clusters through RDMA-based key-value store. In Proceedings of the 2015 44th International Conference on Parallel Processing (ICPP), Beijing, China, 1–4 September 2015; pp. 280–289. [Google Scholar]
- Cui, Y.; Liao, X.; Peng, S.; Lu, Y.; Yang, C.; Wang, B.; Wu, C. Large-scale neo-heterogeneous programming and optimization of SNP detection on Tianhe-2. In Proceedings of the International Conference on High Performance Computing, Bangalore, India, 15–20 November 2015; pp. 74–86. [Google Scholar]
- Wu, C.; Schwartz, J.M.; Brabant, G.; Peng, S.; Nenadic, G. Constructing a molecular interaction network for thyroid cancer via large-scale text mining of gene and pathway events. BMC Syst. Biol. 2015, 9 (Suppl 6), S5. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Peng, S.; Lu, Y.; Zhu, W.; Xu, Z.; Zhang, X. mD3DOCKxb: A deep parallel optimized software for molecular docking with Intel Xeon Phi coprocessors. In Proceedings of the IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing, Shenzhen, China, 4–7 May 2015; pp. 725–728. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |


| Test Scenario Name | Hardware Setting | Analysis Setting |
|---|---|---|
| BGI | A Hadoop cluster with 18 nodes. Each node is equipped with 24 cores, 32 GB RAM. The storage uses HDFS, and the total capacity is 12 TB. | 1 master + 17 slaves.Each core is allocated 3 GB RAM, a maximum of 10 cores were used on each node. |
| Orion-1 | A Hadoop cluster initiated and maintained by Orion on Tianhe-2 with 18 nodes. Each node is equipped with 24 cores, 64 GB RAM. Use the Tianhe-2 parallel filesystem directly. | 1 master + 17 slaves.Each core is allocated 8 GB of RAM, a maximum of 6 cores were used on each node. |
| Orion-2 | 1 master + 17 slaves.Each core is allocated 3 GB of RAM, a maximum of 16 cores were used on each node. |
| SOAPGaea Components | BGI | Orion-1 | Orion-2 |
|---|---|---|---|
| FASTQ Filtering | 24 m 43 s | 12 m 50 s | 10 m 23 s |
| Read Alignment | 1 h 35 m 56 s | 48 m 48 s | 49 m 49 s |
| Duplication Removal | 28 m 21s | 15 m 38 s | 9 m 43 s |
| Quality Control | 1 h 30 m 2 s | 1 h 39 m | 46 m 38 s |
| Total processing time | 3 h 59 m 2 s | 2 h 56 m 16 s | 1 h 56 m 33 s |
| Test Scenario Name | Hardware Setting | Analysis Setting |
|---|---|---|
| BGI | A Spark cluster with 18 nodes. Each node is equipped with 24 cores, 32 GB RAM. The storage uses HDFS and the total capacity is 12 TB. | 1 master + 17 slaves. Each core is allocated 3 GB RAM, a maximum of 10 cores were used on each node. |
| Orion-A | A Spark cluster initiated and maintained by Orion on Tianhe-2 with 100 nodes. Each node is equipped with 24 cores, 64 GB RAM. Uses the Tianhe-2 parallel filesystem directly. | 1 master + 99 slaves. 24 cores, a maximum total of 44 GB RAM cores were used on each node. |
| Orion-B | A Spark cluster initiated and maintained by Orion on Tianhe-2 with 250 nodes. Each node is equipped with 24 cores, 64 GB RAM. Uses the Tianhe-2 parallel filesystem directly. | 1 master + 249 slaves. 24 cores, a maximum total of 44 GB RAM cores were used on each node. |
| GaeaDuplicate_Spark | Read In | Compute | Write Out | Total |
|---|---|---|---|---|
| BGI | 17 m | 1.1 h | 40 m | 2 h |
| Orion-A | 25 m | 14 m | 40 m | 1.3 h |
| Orion-B | 32 m | 6 m | 25 m | 1.1 h |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Wu, C.; Lu, K.; Fang, L.; Zhang, Y.; Li, S.; Guo, G.; Du, Y. An Interface for Biomedical Big Data Processing on the Tianhe-2 Supercomputer. Molecules 2017, 22, 2116. https://doi.org/10.3390/molecules22122116
Yang X, Wu C, Lu K, Fang L, Zhang Y, Li S, Guo G, Du Y. An Interface for Biomedical Big Data Processing on the Tianhe-2 Supercomputer. Molecules. 2017; 22(12):2116. https://doi.org/10.3390/molecules22122116
Chicago/Turabian StyleYang, Xi, Chengkun Wu, Kai Lu, Lin Fang, Yong Zhang, Shengkang Li, Guixin Guo, and YunFei Du. 2017. "An Interface for Biomedical Big Data Processing on the Tianhe-2 Supercomputer" Molecules 22, no. 12: 2116. https://doi.org/10.3390/molecules22122116




