The Complete Amomum kravanh Chloroplast Genome Sequence and Phylogenetic Analysis of the Commelinids
Abstract
:1. Introduction
2. Results and Discussion
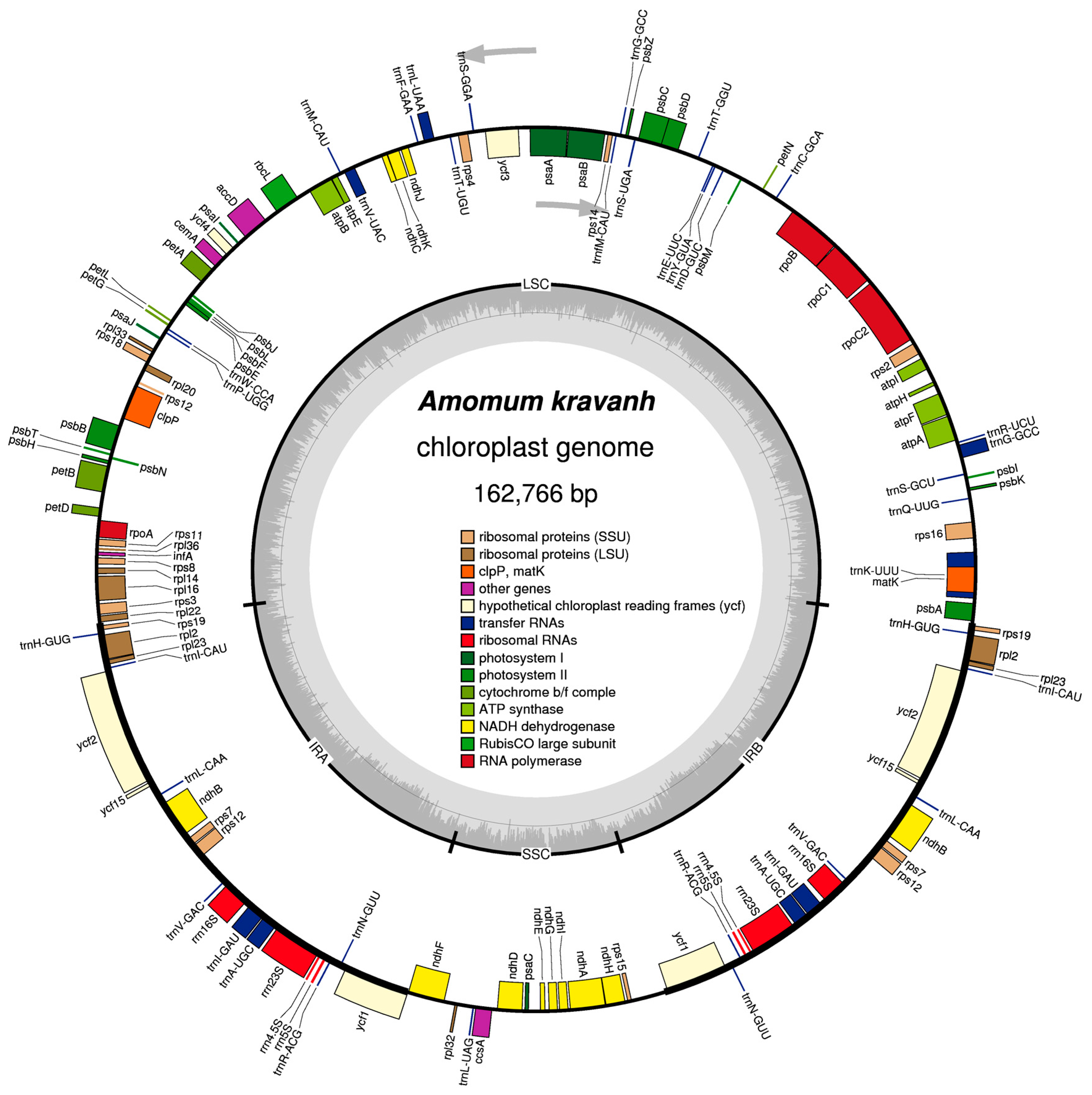
2.1. Characteristics of the A. kravanh cp Genome
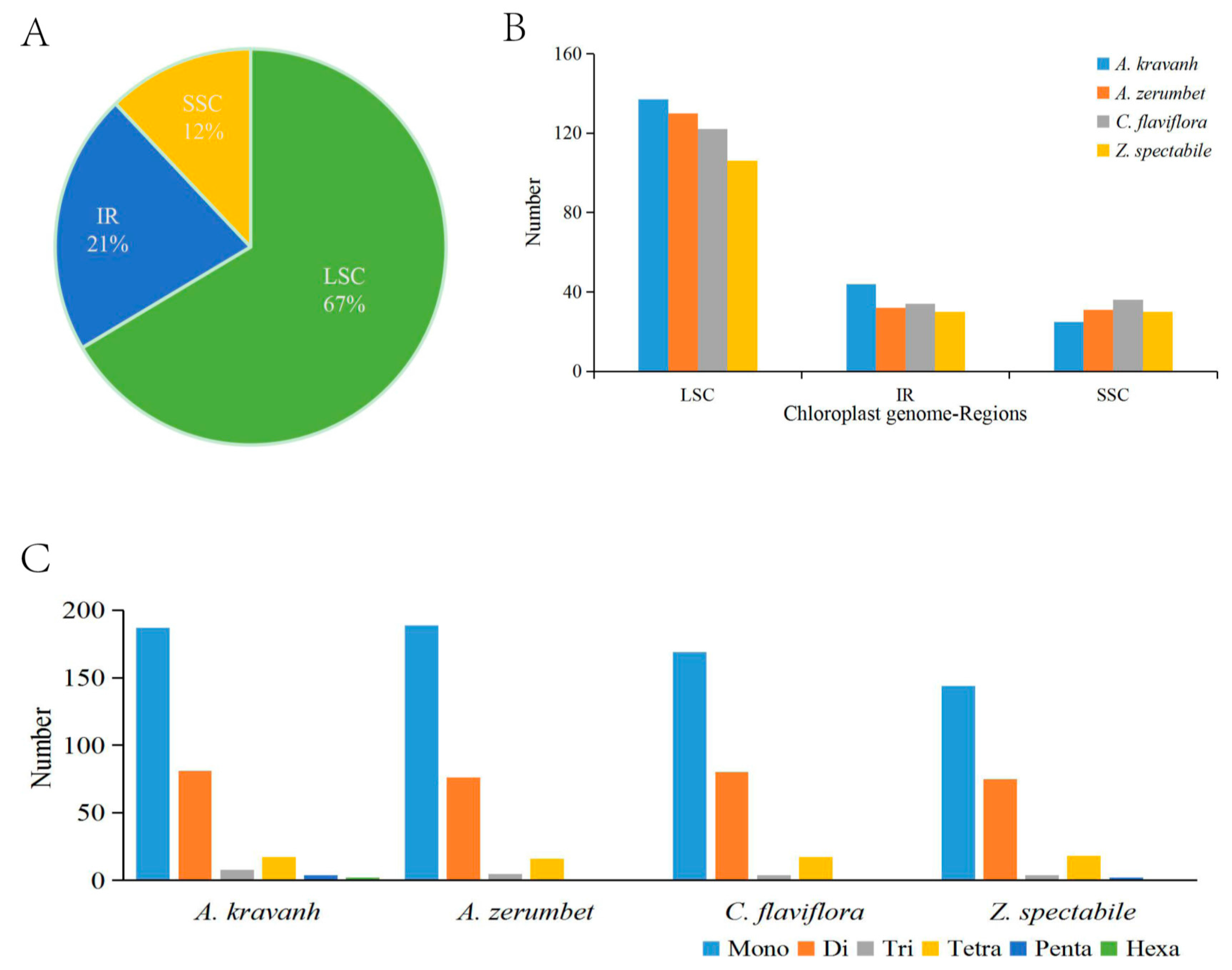
2.2. Analysis of SSRs and Long Repeats
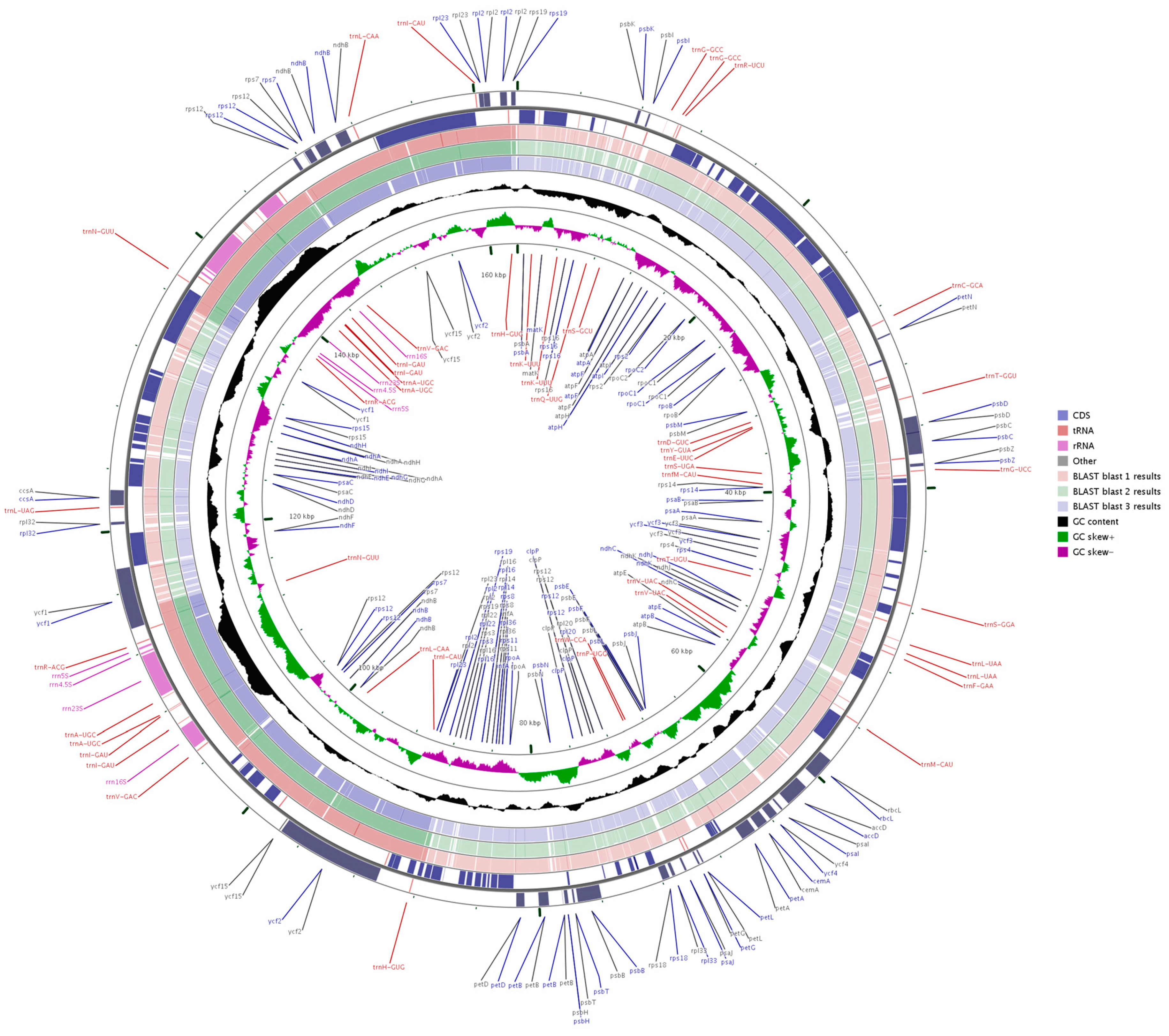
2.3. Comparative Chloroplast Genomic Analysis
2.4. IR Contraction and Expansion
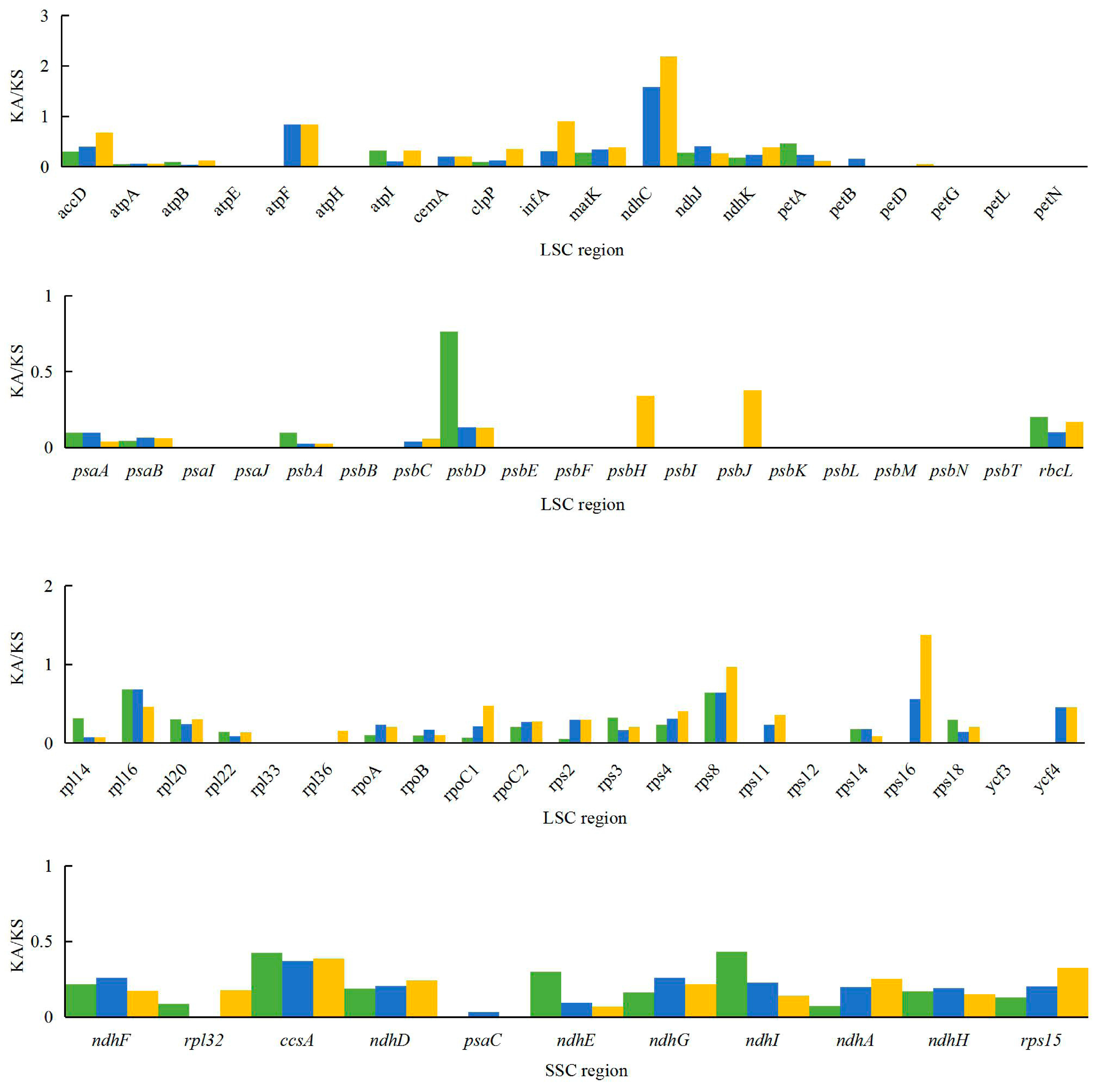
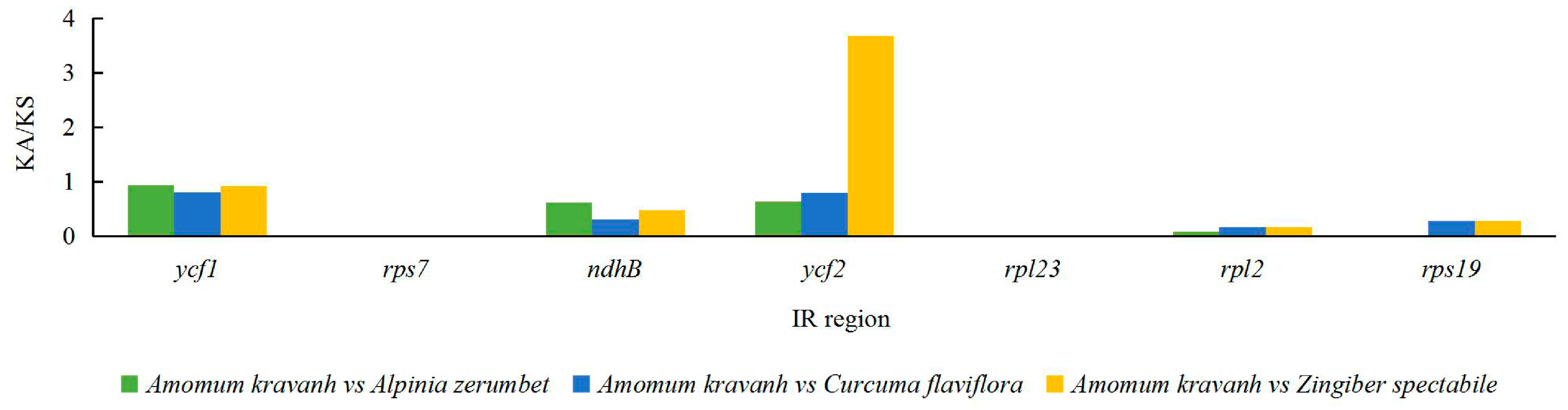
2.5. Analysis of Synonymous and Non-Synonymous Substitution Rates
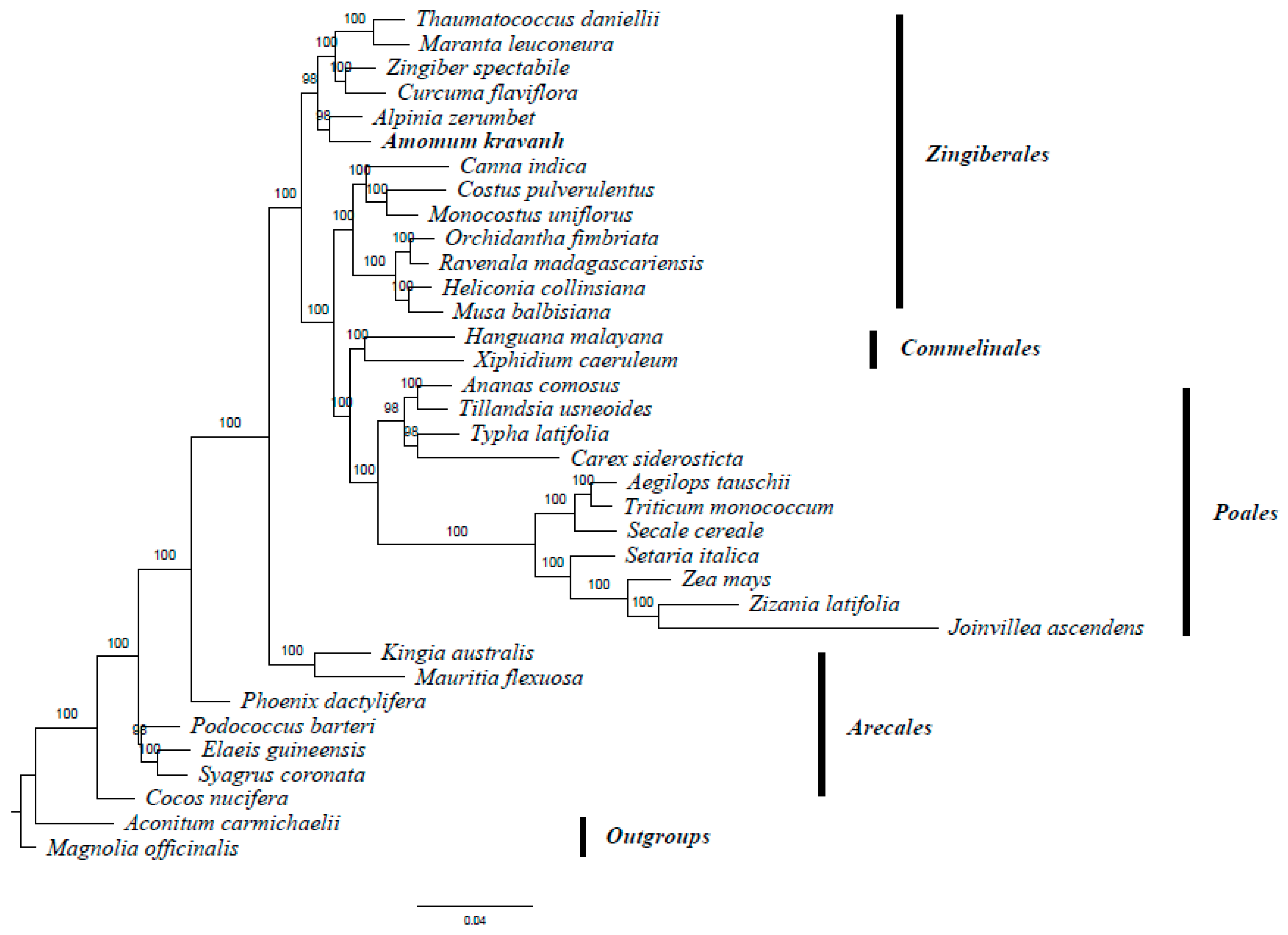
2.6. Phylogenetic Analysis
3. Materials and Methods
3.1. DNA Sequencing and Genome Assembly
3.2. Genome Annotation
3.3. Sequence Analyses
3.4. Genome Comparison
3.5. Analysis of Synonymous and Non-Synonymous Substitution Rates
3.6. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shu, Y.M.; Zhou, M.Q.; Zhang, Y.J.; Yue, W.Q. Clinical efficacy study of Yishen\Jiangzhuo granule on chronic renal failure. Chin. Tradit. Pat. Med. 1993, 2, 20–21. [Google Scholar]
- Pu, Q.; Qin, H.Z.; Huang, Y.Q.; Xie, P.; Luo, J.; Wong, M.Z. Advances in studies on the correlation between medicinal properties and genetic relationship of five genera in Zingiberaceae. China Pharm. 2016, 27, 3301–3303. [Google Scholar]
- Luo, Y.; Lu, L.; Wortley, A.H.; Li, D.Z.; Wang, H.; Blackmore, S. Evolution of Angiosperm Pollen. 3. Monocots1. Ann. Mo. Bot. Gard. 2015, 101, 406–455. [Google Scholar] [CrossRef]
- Nie, X.; Lv, S.; Zhang, Y.; Du, X.H.; Wang, L.; Biradar, S.S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Qian, J.; Sun, Z.Y.; Xu, X.L.; Chen, S.L. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; Li, X.; Qian, J.; Wang, L.; Ma, L.; Tian, X. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules 2016, 21, 1029–1031. [Google Scholar] [CrossRef] [PubMed]
- Clegg, M.T.; Gaut, B.S.; Learn, G.H.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.Y.; Wu, H.; Wang, Y.L.; Gao, L.M.; Zhang, S.Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.K.; Lee, H.L.; Sun, B.Y.; Chung, M.Y.; Kim, K.J. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae); comparative evolutionary analyses with other three asterids. Mol. Ceflls. 2012, 33, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.J.; McNicol, J.W.; Meyer, R.C.; Ramsay, G.; De Jong, W.S. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor. Appl. Genet. 1999, 99, 859–867. [Google Scholar] [CrossRef]
- Provan, J. Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana. Mol. Ecol. 2000, 9, 2183–2185. [Google Scholar] [CrossRef] [PubMed]
- Flannery, M.L.; Mitchell, F.J.; Coyne, S.; Kavanagh, T.A.; Burke, J.I.; Salamin, N. Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 2006, 113, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.H.; Zhao, Z.L.; Xu, H.X.; Chen, S.L.; Dorje, G. The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion. Gene 2016, 577, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.J.; Cheng, C.L.; Chang, C.C.; Wu, C.L.; Su, T.M.; Chaw, S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evolut. Biol. 2008, 8, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Ye, Y.; Bai, T.; Xu, M.; Xu, L.-A. Complete chloroplast genome of Pinus massoniana (Pinaceae): Gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion. Molecules 2017, 22, 1528. [Google Scholar] [CrossRef] [PubMed]
- De Cambiaire, J.C.; Otis, C.; Lemieux, C.; Turmel, M. The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evolut. Biol. 2006, 6, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, B.; Zhu, W.; Sun, L.; Tian, J.; Wang, X. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene 2013, 528, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Drescher, A.; Ruf, S.; Calsa, T.J.; Carrer, H.; Bock, R. The two largest chloroplast genome-encoded open reading frames of higher plant are essential genes. Plant J. 2000, 22, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Johansen, L.B.; Hoot, S.B. Phylogeny of Orchidantha (Lowiaceae) and the Zingiberales based on six DNA regions. Syst. Botany 2005, 30, 106–117. [Google Scholar] [CrossRef]
- Givnish, T.J.; Evans, T.M.; Pires, J.C.; Systma, K.J. Polyphyly and convergent morphological evolution in Commelinales and Commelinidae: Evidence from rbcL sequence data. Mol. Phylogenet. Evolut. 1999, 12, 360–385. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.B. Phylogenetic analysis of the subfamily Alpinioideae (Zingiberaceae), particularly Etlingera giseke, based on nuclear and plastid DNA. Plant Syst. Evolut. 2004, 245, 239–258. [Google Scholar] [CrossRef]
- Smith, J.F.; Kress, W.J.; Zimmer, E.A. Phylogenetic analysis of the Zingiberales based on rbcL sequences. Ann. Mo. Bot. Gard. 1993, 80, 620–630. [Google Scholar] [CrossRef]
- Dong, H.; Mei, Q.C.; Xu, G.J.; Xu, L.S. The herbal textual research of Alpinia katsumadai and cardamom. J. Chin. Mater. Med. 1992, 17, 451–453. [Google Scholar]
- Li, X.W.; Hu, Z.G.; Lin, X.H.; Li, Q.; Gao, H.H.; Luo, G.A. High-throughput pyrosequencing of the complete chloroplast genome of Magnolia officinalis and its application in species identification. Acta Pharm. Sin. 2012, 47, 124–130. [Google Scholar]
- Liu, C.; Shi, Y.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.E.; Searle, S.M.; Harris, N.; Gibson, M.; Iyer, V.; Richter, J. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, RESEARCH0082. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellar Genome DRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evolut. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.M.; Sun, J.T.; Xue, X.F.; Zhu, W.C.; Hong, X.Y. Development and characterization of 18 Novel EST-SSRs from the western flower Thrips, Frankliniella occidentalis (Pergande). Int. J. Mol. Sci. 2012, 13, 2863–2876. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evolut. 2008, 25, 1253. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. In Proceedings of the Teragrid Conference: Extreme Digital Discovery, Salt Lake City, UT, USA, 18–21 July 2011. [Google Scholar]
Sample Availability: Sequence data of Amomum kravanh are available from the authors. |

| T (U) (%) | C (%) | A (%) | G (%) | Length (bp) | ||
|---|---|---|---|---|---|---|
| LSC | 33.8 | 17.3 | 32.3 | 16.6 | 87,728 | |
| SSC | 34.2 | 15.6 | 36.0 | 14.2 | 15,390 | |
| IRA | 28.8 | 19.8 | 30.1 | 21.3 | 29,824 | |
| IRB | 30.1 | 21.3 | 28.8 | 19.8 | 29,824 | |
| Total | 32.2 | 18.3 | 31.6 | 17.8 | 162,766 | |
| CDS | 31.6 | 17.2 | 31.3 | 19.9 | 81,642 | |
| 1st position | 24.0 | 18.3 | 31.2 | 26.6 | 27,214 | |
| 2nd position | 32.0 | 20.2 | 29.9 | 17.5 | 27,214 | |
| 3rd position | 38.0 | 13.2 | 32.8 | 15.5 | 27,214 |
| Amino Acid | Codon | Count | RSCU | tRNA | Amino Acid | Codon | Count | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 997 | 1.29 | Tyr | UAU | 818 | 1.58 | ||
| Phe | UUC | 545 | 0.71 | trnF-GAA | Tyr | UAC | 219 | 0.42 | trnY-GUA |
| Leu | UUA | 900 | 1.93 | trnL-UAA | Stop | UAA | 49 | 1.69 | |
| Leu | UUG | 579 | 1.24 | trnL-CAA | Stop | UAG | 21 | 0.72 | |
| Leu | CUU | 586 | 1.25 | His | CAU | 526 | 1.6 | ||
| Leu | CUC | 192 | 0.41 | His | CAC | 132 | 0.4 | trnH-GUG | |
| Leu | CUA | 389 | 0.83 | trnL-UAG | Gln | CAA | 725 | 1.54 | trnQ-UUG |
| Leu | CUG | 159 | 0.34 | Gln | CAG | 218 | 0.46 | ||
| Ile | AUU | 1168 | 1.47 | Asn | AAU | 1010 | 1.54 | ||
| Ile | AUC | 438 | 0.55 | trnI-GAU | Asn | AAC | 298 | 0.46 | trnN-GUU |
| Ile | AUA | 772 | 0.97 | trnI-CAU | Lys | AAA | 1105 | 1.47 | trnK-UUU |
| Met | AUG | 636 | 1 | trn(f)M-CAU | Lys | AAG | 399 | 0.53 | |
| Val | GUU | 535 | 1.45 | Asp | GAU | 896 | 1.64 | ||
| Val | GUC | 168 | 0.46 | trnV-GAC | Asp | GAC | 196 | 0.36 | trnD-GUC |
| Val | GUA | 571 | 1.55 | trnV-UAC | Glu | GAA | 1144 | 1.51 | trnE-UUC |
| Val | GUG | 200 | 0.54 | Glu | GAG | 371 | 0.49 | ||
| Ser | UCU | 622 | 1.74 | Cys | UGU | 240 | 1.55 | ||
| Ser | UCC | 352 | 0.99 | trnS-GGA | Cys | UGC | 69 | 0.45 | trnC-GCA |
| Ser | UCA | 427 | 1.2 | trnS-UGA | Stop | UGA | 17 | 0.59 | |
| Ser | UCG | 196 | 0.55 | Trp | UGG | 469 | 1 | trnW-CCA | |
| Pro | CCU | 454 | 1.64 | Arg | CGU | 372 | 1.35 | trnR-ACG | |
| Pro | CCC | 206 | 0.74 | Arg | CGC | 89 | 0.32 | ||
| Pro | CCA | 328 | 1.18 | trnP-UGG | Arg | CGA | 353 | 1.28 | |
| Pro | CCG | 122 | 0.44 | Arg | CGG | 120 | 0.43 | ||
| Thr | ACU | 546 | 1.57 | Arg | AGA | 545 | 1.97 | trnR-UCU | |
| Thr | ACC | 241 | 0.69 | trnT-GGU | Arg | AGG | 178 | 0.64 | |
| Thr | ACA | 441 | 1.27 | trnT-UGU | Ser | AGU | 436 | 1.22 | |
| Thr | ACG | 161 | 0.46 | Ser | AGC | 108 | 0.3 | trnS-GCU | |
| Ala | GCU | 631 | 1.82 | Gly | GGU | 614 | 1.39 | ||
| Ala | GCC | 202 | 0.58 | Gly | GGC | 144 | 0.33 | trnG-GCC | |
| Ala | GCA | 440 | 1.27 | trnA-UGC | Gly | GGA | 729 | 1.65 | |
| Ala | GCG | 115 | 0.33 | Gly | GGG | 285 | 0.64 |
| No. | Size (bp) | Type | Repeat 1 Start | Repeat 1 Location | Repeat 2 Start | Repeat 2 Location | Region | E-Value |
|---|---|---|---|---|---|---|---|---|
| 1 | 30 | C | 9027 | IGS | 53,756 | IGS | LSC | 7.08 × 10−4 |
| 2 | 30 | F | 10,500 | trnG-GCC | 38,870 | trnG-UCC | LSC | 7.08 × 10−4 |
| 3 | 30 | F | 32,612 | IGS | 32,641 | IGS | LSC | 7.08 × 10−4 |
| 4 | 30 | F | 33,321 | IGS | 133,080 | IGS | LSC; IRa | 7.08 × 10−4 |
| 5 | 58 | F | 41,076 | psaB (CDS) | 43,300 | psaA (CDS) | LSC | 7.47 × 10−20 |
| 6 | 37 | F | 41,120 | psaB (CDS) | 43,344 | psaA (CDS) | LSC | 2.36 × 10−9 |
| 7 | 30 | F | 70,833 | IGS | 70,860 | IGS | LSC | 6.46 × 10−9 |
| 8 | 42 | F | 71,482 | rps18 (CDS) | 71,503 | rps18 (CDS) | LSC | 1.19 × 10−10 |
| 9 | 46 | F | 90,440 | trnI-CAU | 90,490 | IGS | IRb | 2.08 × 10−16 |
| 10 | 30 | F | 93,108 | ycf2 (CDS) | 93,129 | ycf2 (CDS) | IRb | 7.08 × 10−4 |
| 11 | 30 | F | 157,337 | ycf2 (CDS) | 157,358 | ycf2 (CDS) | IRa | 7.08 × 10−4 |
| 12 | 46 | F | 159,958 | IGS | 160,008 | IGS | IRa | 2.08 × 10−16 |
| 13 | 30 | F | 159,977 | IGS | 160,027 | IGS | IRa | 7.08 × 10−4 |
| 14 | 34 | P | 3990 | trnK-UUU | 3996 | trnK-UUU | LSC | 4.08 × 10−6 |
| 15 | 31 | P | 8746 | IGS | 47,581 | trnS-GGA | LSC | 1.96 × 10−4 |
| 16 | 32 | P | 30,943 | IGS | 30,973 | IGS | LSC | 5.41 × 10−5 |
| 17 | 30 | P | 33,321 | IGS | 117,383 | ycf1 (CDS) | LSC; IRb | 7.08 × 10−4 |
| 18 | 30 | P | 34,662 | IGS | 62,566 | IGS | LSC | 7.08 × 10−4 |
| 19 | 32 | P | 39,185 | IGS | 39,226 | IGS | LSC | 4.04 × 10−10 |
| 20 | 31 | P | 67,014 | IGS | 67,069 | IGS | LSC | 6.76 × 10−6 |
| 21 | 46 | P | 90,440 | trnI-CAU | 159,958 | IGS | IRb; IRa | 2.08 × 10−16 |
| 22 | 46 | P | 90,490 | IGS | 160,008 | IGS | IRb; IRa | 2.08 × 10−16 |
| 23 | 30 | P | 93,108 | ycf2 (CDS) | 157,335 | ycf2 (CDS) | IRb; IRa | 7.08 × 10−4 |
| 24 | 30 | P | 93,129 | ycf2 (CDS) | 157,356 | ycf2 (CDS) | IRb; IRa | 7.08 × 10−4 |
| 25 | 181 | P | 117,370 | ycf1 (CDS) | 132,942 | ycf1 (CDS) | IRb; IRa | 7.93 × 10−100 |
| 26 | 31 | R | 13,116 | atpF (intron) | 34,886 | IGS | LSC | 6.76 × 10−6 |
| 27 | 34 | R | 33,365 | IGS | 33,374 | IGS | LSC | 4.08 × 10−6 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Li, Q.; Hu, Z.; Li, X.; Chen, S. The Complete Amomum kravanh Chloroplast Genome Sequence and Phylogenetic Analysis of the Commelinids. Molecules 2017, 22, 1875. https://doi.org/10.3390/molecules22111875
Wu M, Li Q, Hu Z, Li X, Chen S. The Complete Amomum kravanh Chloroplast Genome Sequence and Phylogenetic Analysis of the Commelinids. Molecules. 2017; 22(11):1875. https://doi.org/10.3390/molecules22111875
Chicago/Turabian StyleWu, Mingli, Qing Li, Zhigang Hu, Xiwen Li, and Shilin Chen. 2017. "The Complete Amomum kravanh Chloroplast Genome Sequence and Phylogenetic Analysis of the Commelinids" Molecules 22, no. 11: 1875. https://doi.org/10.3390/molecules22111875




