Digital Gene Expression Profiling to Explore Differentially Expressed Genes Associated with Terpenoid Biosynthesis during Fruit Development in Litsea cubeba
Abstract
:1. Introduction
2. Results
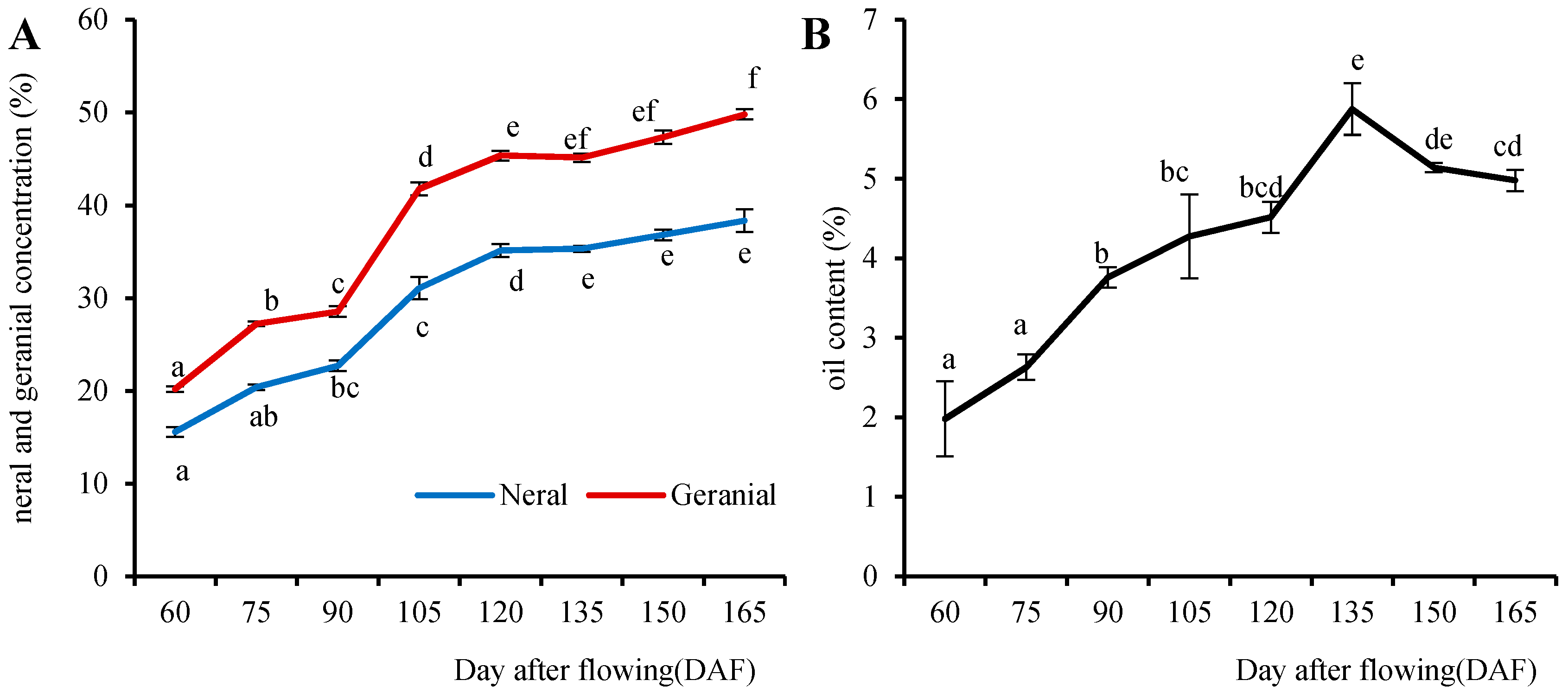
2.1. Accumulation of Citral Concentrations at Different Stages of Fruit Development
2.2. Analysis of DGE Libraries and Mapping
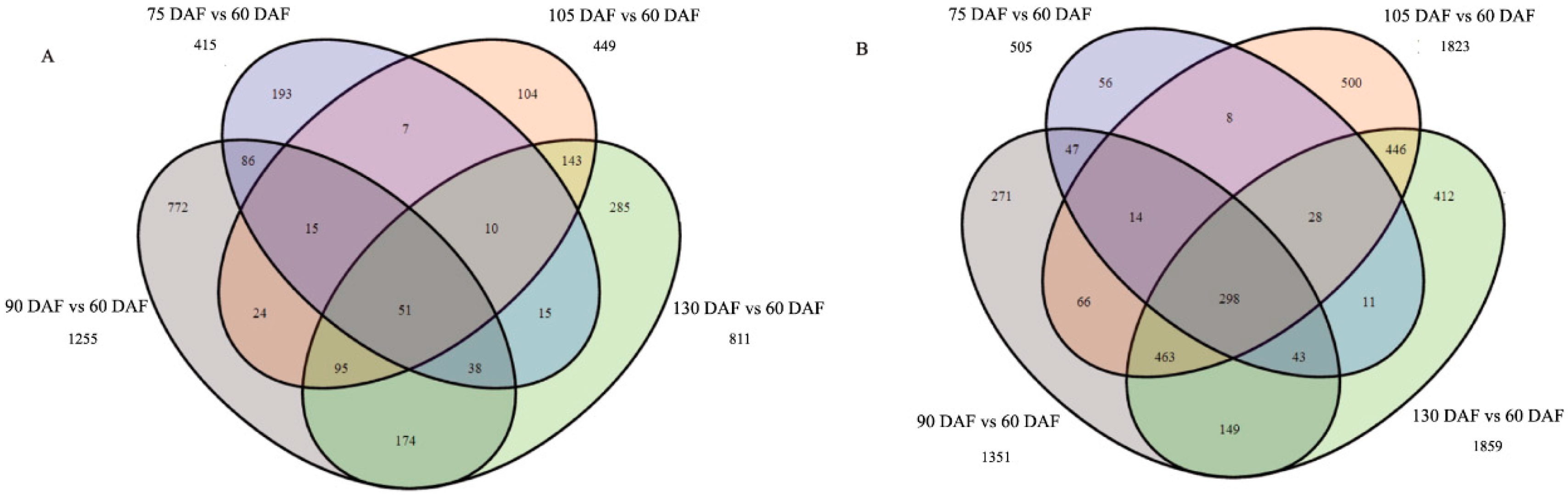
2.3. Differential Gene Expression Analysis
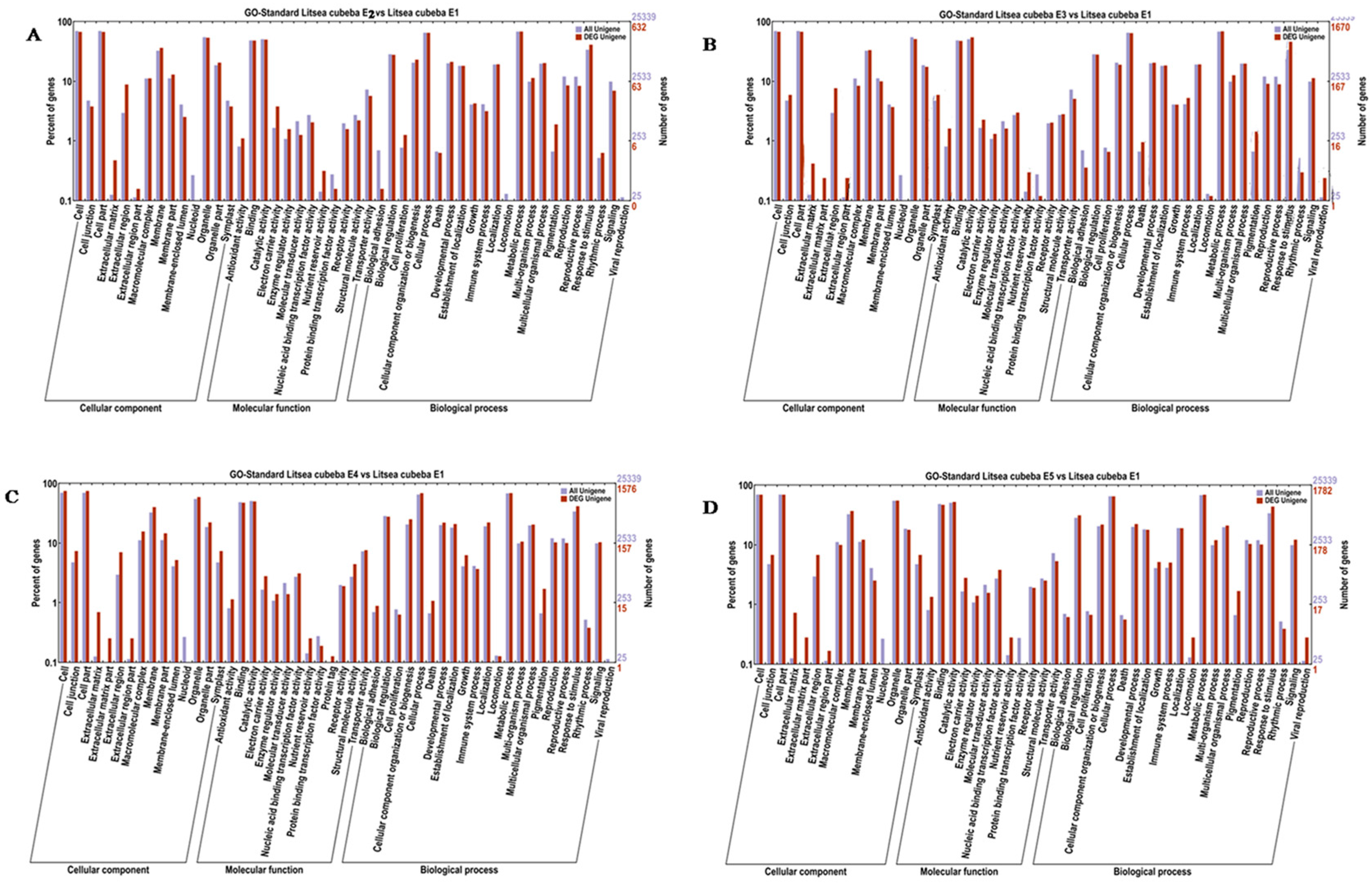
2.4. GO Functional Enrichment Analysis of the DEGs
2.5. Pathway Enrichment Analysis of DEGs Using KEGG
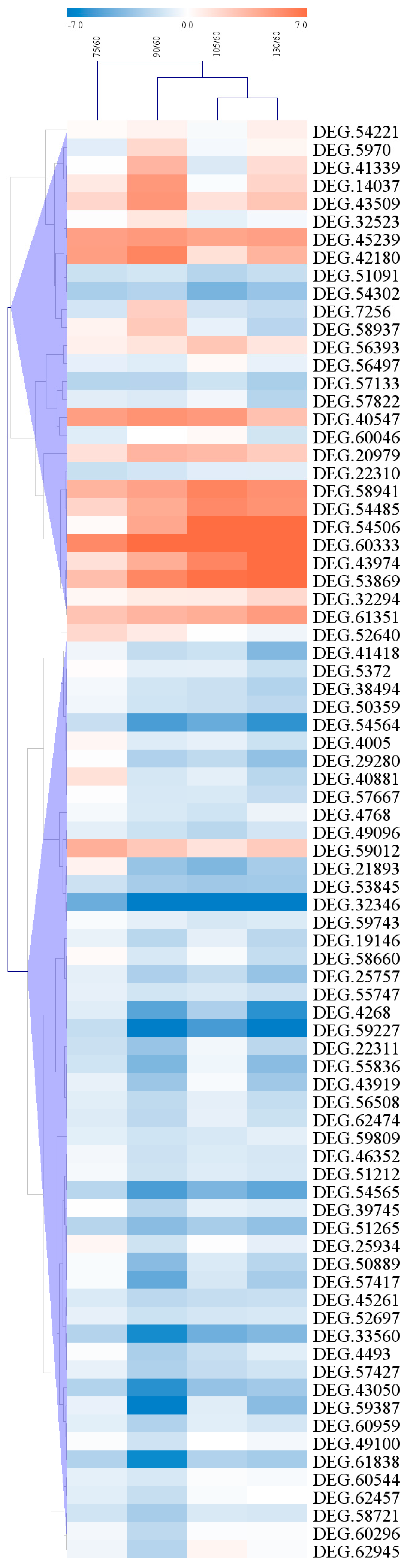
2.6. Clustering of Candidate Genes
2.7. Confirmation of Differential Genes Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement and Identification of Essential Oil Compositions
4.3. Total RNA Extraction, cDNA Library Construction, and High Throughput Sequencing
4.4. Sequencing Data and Differentially Expressed Gene Analysis
4.5. Functional and Clustering Analysis of the Differentially Expressed Genes
4.6. Quantitative Real-Time PCR Validation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, Y.; Wang, Y.; Han, X.; Si, L.; Wu, Q.; Lin, L. Biology and chemistry of Litsea cubeba, a promising industrial tree in China. J. Essent. Oil Res. 2013. [Google Scholar] [CrossRef]
- Han, X.; Wang, Y.; Chen, Y.; Lin, L.; Wu, Q. Transcriptome sequencing and expression analysis of terpenoid biosynthesis genes in Litsea cubeba. PLoS ONE 2013, 8, e76890. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Singh, U.; Meli, V.; Kumar, V.; Kumar, A.; Irfan, M.; Chakraborty, N.; Chakraborty, S.; Datta, A. Induction of senescence and identification of differentially expressed genes in Tomato in response to monoterpene. PLoS ONE 2013, 8, e76029. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Lou, Y; Mao, Y.; Lu, S.; Wang, L.; Chen, X. Plant terpenoids: biosynthesis and ecological Functions. J. Integr. Plant Biol. 2007, 49, 179–186. [Google Scholar] [CrossRef]
- Nagegowda, D. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010, 584, 2965–2973. [Google Scholar] [CrossRef] [PubMed]
- Vollack, K.; Bach, T. Cloning of a cDNA encoding cytosolic acetoacetyl-coenzyme A thiolase from radish by functional expression in Saccharomyces cerevisiae. Plant Physiol. 1996, 111, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Montamat, F.; Guilloton, M.; Karst, F.; Delrot, S. Isolation and characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-coenzyme A synthase. Gene 1995, 167, 197–201. [Google Scholar] [CrossRef]
- Nagegowda, D.; Bach, T.; Chye, M. Brassica juncea HMG-CoA synthase 1: expression and characterization of recombinant wild-type and mutant enzymes. Biochem. J. 2004, 383, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Sirinupong, N.; Suwanmanee, P.; Doolittle, R.F.; Suvachitanont, W. Molecular cloning of a new cDNA and expression of 3-hydroxy-3-methylglutaryl-CoA synthase gene from Hevea brasiliensis. Planta 2005, 221, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.; Rhodes, D.; Dudareva, N. The role of methyl-erythritol-phosphate pathway in rhythmic emission of volatiles. In The Chloroplast: Basics and Application; Rebeiz, C.A., Benning, C., Daniel, H., Eds.; Springer Press: Amsterdam, The Netherlands, 2010; Volume 31, pp. 139–153. [Google Scholar]
- Rodríguez-Concepción, M.; Ahumada, I.; Diez-Juez, E.; Sauret-Güeto, S.; Lois, L.; Gallego, F.; Carretero-Paulet, L.; Campos, N.; Boronat, A. 1-Deoxy-d-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 2001, 27, 213–222. [Google Scholar]
- Chang, Y.; Chu, F. Molecular cloning and characterization of monoterpene synthases from Litsea cubeba (Lour.) Persoon. Tree Genet. Genomes 2011, 7, 835–844. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Q.; He, G.; Wang, Y.; Yang, S.; Chen, Y.; Gao, M. Cloning and SNP analysis of 1-Deoxy-d-xylulose-5-phosphate Reductoisomerases (DXR) in Litsea cubeba. For. Res. 2015, 28, 93–100. [Google Scholar]
- Eveland, A.L.; Satoh-Nagasawa, N.; Goldshmidt, A.; Meyer, S.; Beatty, M.; Sakai, H.; Ware, D.; Jackson, D. Digital Gene Expression Signatures for Maize Development. Plant Physiol. 2010, 154, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, F.; Dong, Y.; Wang, N.; Sun, Y.; Li, X.; Liu, L.; Fan, X.; Yin, H.; Jing, Y. Sequence mining and transcript profiling to explore differentially expressed genes associated with lipid biosynthesis during soybean seed development. BMC Plant Biol. 2012, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, Y.; Zhu, B.; Fang, T.; Fang, Y.; Wang, Y. Digital gene expression analysis of gene expression differences within Brassica diploids and allopolyploids. BMC Plant Biol. 2015, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- AC'tHoen, P.; Ariyurek, Y.; Thygesen, H.H.; Vreugdenhil, E.; Vossen, R.H.; de Menezes, R.X.; Boer, J.M.; van Ommen, G.J.B.; den Dunnen, J.T. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008, 36, e141. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Li, J.; Schmidt-Küntzel, A.; Warren, W.C.; Barsh, G.S. Digital gene expression for non-model organisms. Genome Res. 2011, 21, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Song, M.; Fan, S.; Yu, S. Transcriptomic analysis of differentially expressed genes during anther development in genetic male sterile and wild type cotton by digital gene-expression profiling. BMC Genom. 2013, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, X.; Liao, X.; Zuo, T.; Qin, C.; Cao, S.; Dong, L.; Zhou, H.; Zhang, Y.; Liu, S. Genome-wide comparative analysis of digital gene expression tag profiles during maize ear development. Genomics 2015, 106, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Liu, S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Arasu, M.; Al-Dhabi, N.; Lim, S.; Kin, Y.; Lee, S.; Park, S. Expression of terpenoid biosynthetic genes and accumulation of chemical constituents in Valeriana fauriei. Molecules 2016, 21, 691. [Google Scholar] [CrossRef] [PubMed]
- Gillissen, B.; Bürkle, L.; André, B.; Kühn, C.; Rentsch, D.; Brandl, B.; Frommer, W.B. A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in arabidopsis. Plant Cell 2000, 12, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Thwe, A.A.; Li, X.; Tuan, P.A.; Lee, S.; Lee, J.W.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.U. Accumulation of astragalosides and related gene expression in different organs of Astragalus membranaceus BGE. Var mongholicus (BGE.). Molecules 2014, 19, 10922–10935. [Google Scholar] [CrossRef] [PubMed]
- Lykkesfeldt, J.; Moller, B. Synthesis of benzylglucosinolate in Tropaeolum majus L. (isothiocyanates as potent enzyme inhibitors). Plant Physiol. 1993, 102, 609–613. [Google Scholar] [PubMed]
- Penfold, A.R.; Morrison, F.R.; Willis, J.L.; McKern, H.H.G.; Spies, M.C. The essential oil of a physiological form of Eucalyptus citriodora Hook. J. Proc. R. Soc. N. S. W. 1951, 85, 120–122. [Google Scholar]
- Iijima, Y.; Koeduka, T.; Suzuki, H.; Kubota, K. Biosynthesis of geranial, a potent aroma compound in ginger rhizome (Zingiber officinale): Molecular cloning and characterization of geraniol dehydrogenase. Plant Biotechnol. 2014, 31, 525–534. [Google Scholar] [CrossRef]
- Kimura, K.; Iwata, I.; Nishimura, H. Relationship between Acid-Catalyzed Cyclization of Citral and Deterioration of Lemon Flavor. Agric. Biol. Chem. 1982, 46, 1387–1389. [Google Scholar] [CrossRef]
- Wolken, W.A.M.; ten Have, R.; van der Werf, M.J. Amino Acid-Catalyzed Conversion of Citral: cis−trans Isomerization and Its Conversion into 6-Methyl-5-hepten-2-one and Acetaldehyde. J. Agric. Food Chem. 2000, 48, 5401–5405. [Google Scholar] [CrossRef] [PubMed]
- Iijima, Y.; Wang, G.; Fridman, E.; Pichersky, E. Analysis of the enzymatic formation of citral in the glands of sweet basil. Arch. Biochem. Biophys. 2006, 448, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Sato-Masumoto, N.; Ito, M. Two types of alcohol dehydrogenase from Perilla can form citral and perillaldehyde. Phytochemistry 2014, 104, 12–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, T.; Endo, T.; Fujii, H.; Hara, M.; Ueda, T.; Kita, M.; Omura, M. Molecular cloning and functional characterization of four monoterpene synthase genes from Citrus unshiu Marc. Plant Sci. 2004, 166, 49–58. [Google Scholar] [CrossRef]
- Wang, G.; Tian, L.; Aziz, N.; Broun, P.; Dai, X.; He, J.; King, A.; Zhao, P.X.; Dixon, R.A. Terpene Biosynthesis in Glandular Trichomes of Hop. Plant Physiol. 2008, 148, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, M.; Naghavi, M.R.; Alizadeh, H.; Soltanloo, H. Expression of artemisinin biosynthesis genes in eight Artemisia species at three developmental stages. Ind. Crops Prod. 2015, 76, 836–843. [Google Scholar] [CrossRef]
- Bouvier, F.; Suire, C.; D’Harlingue, A.; Backhaus, R.; Camara, B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 2000, 24, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Van Schie, C.; Ament, K.; Schmidt, A.; Lange, T.; Haring, M.; Schuurink, R. Geranyl diphosphate synthase is required for biosynthesis of gibberellins. Plant J. 2007, 52, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Hsieh, F.; Ko, T.; Teng, K.; Liang, P.; Wang, A. Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation. Plant Cell 2010, 22, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Poudereux, I.; Kutzner, E.; Huber, C.; Segura, J.; Eisenreich, W.; Arrillaga, I. Metabolic cross-talk between pathways of terpenoid backbone biosynthesis in spike lavender. Plant Physiol. Biochem. 2015, 95, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yao, L. Cloning and expression profile of 1-Deoxy-d-Xylulose 5-phosphate reductoisomerase gene from an oil-bearing rose. Russ. J. Plant Physiol. 2014, 61, 548–555. [Google Scholar] [CrossRef]
- Jordan, M.J.; Margaria, C.A.; Shaw, P.E.; Goodner, K.L. Aroma active components in aqueous kiwi fruit essence and kiwi fruit puree by GC-MS and multidimensional GC/GC-O. J. Agric. Food Chem. 2002, 50, 5386–5390. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, Y.; Du, M.; Zhang, J. Characterization of the volatiles and active components in ethanol extracts of fruits of Litsea cubeba (Lour.) by gas chromatography-mass spectrometry (GC-MS) and gas chromatography-olfactometry (GC-O). J. Med. Plants Res. 2011, 5, 3298–3303. [Google Scholar]
- Kent, W.J. BLAT--the BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from rna-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Romualdi, C.; Bortoluzzi, S.; D’Alessi, F.; Danieli, G.A. IDEG6: A web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol. Genom. 2003, 12, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Han, X.; Chen, Y.; Wu, Q.; Wang, Y.D. Identification of appropriate reference genes for normalizing transcript expression by quantitative real-time PCR in Litsea cubeba. Mol. Genet. Genom. 2013, 288, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
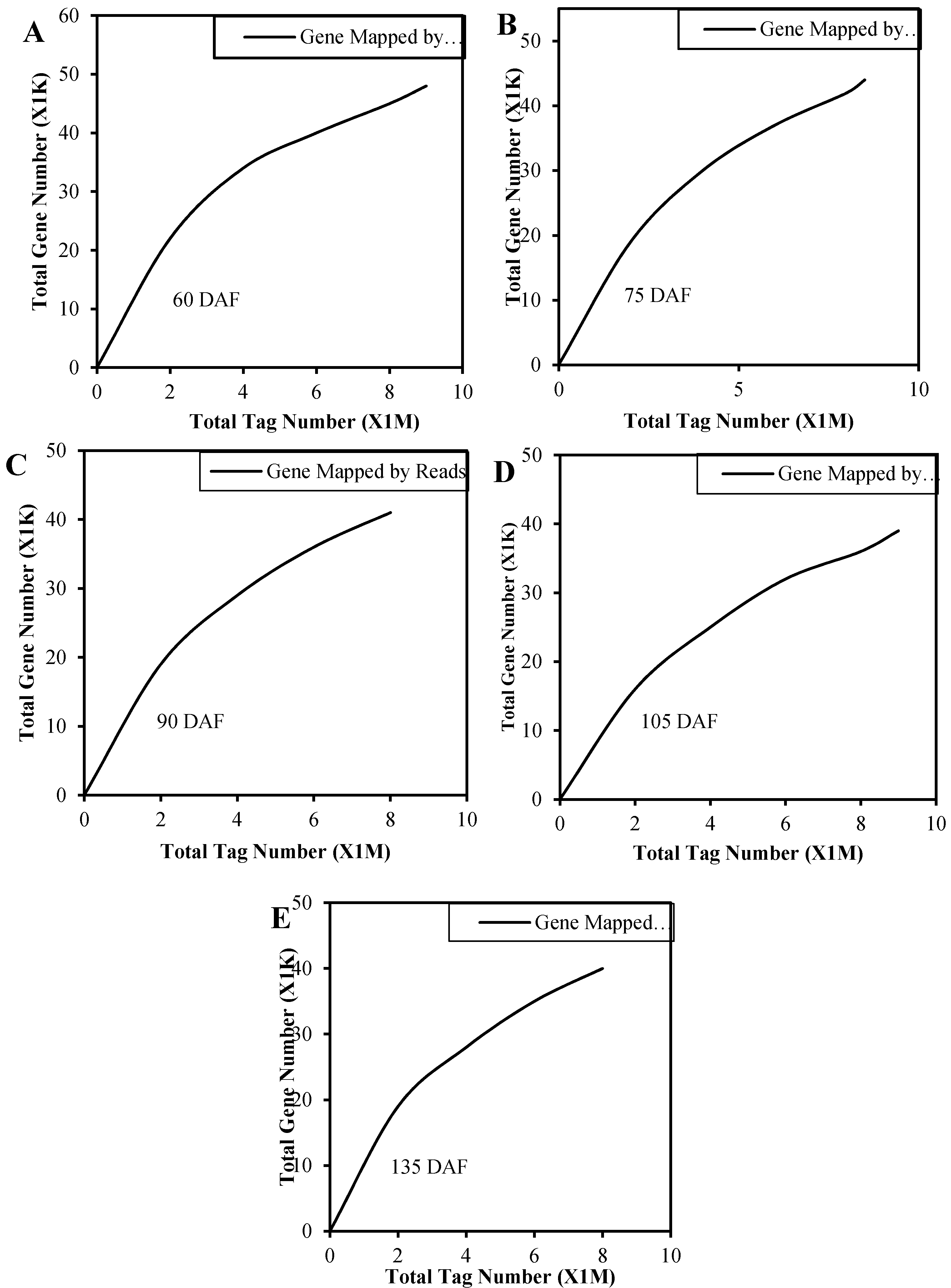

| Samples | Read Number | GC Percentage (%) | Total Mapped Reads 1 (Mapped Ratio %) | Uniquely Mapped Reads 2 (Mapped Ratio %) |
|---|---|---|---|---|
| 60 DAF | 8,971, 349 | 46.38 | 7,845,525 (87.45) | 5,707,386 (63.62) |
| 75 DAF | 8,196,825 | 47.57 | 7,349,267 (89.66) | 5,410,062 (66.00) |
| 90 DAF | 8,531,708 | 47.26 | 7,557,873 (88.58) | 5,483,701 (64.27) |
| 105 DAF | 9,268,559 | 47.50 | 8,387,997 (90.50) | 5,827,196 (62.87) |
| 130 DAF | 8,194,664 | 47.31 | 7,346,494 (89.64) | 5,122,138 (62.50) |
| Gene ID | KEGG Annotation | log2 Ratio 1 | |||
|---|---|---|---|---|---|
| 75 DAF | 90 DAF | 105 DAF | 135 DAF | ||
| 59743 | acetyl-CoA C-acetyltransferase | −0.18057 | −0.86507 | −1.3505 | −1.14886 |
| 4005(HMGS) | hydroxymethylglutaryl-CoA synthase | 0.405639 | −1.05733 | −0.74322 | −1.62293 |
| 57667(HMGS) | hydroxymethylglutaryl-CoA synthase | −0.03242 | −1.2824 | −1.24393 | −1.90689 |
| 43919(HMGR) | hydroxymethylglutaryl-CoA reductase (NADPH) | −0.72437 | −3.24793 | −0.20353 | −3.24793 |
| 60046(HMGR) | hydroxymethylglutaryl-CoA reductase (NADPH) | −1.01749 | 0.017278 | 0.268817 | −1.46815 |
| 50889(IDI) | isopentenyl-diphosphate delta-isomerase | −0.16506 | −3.79442 | −1.15056 | −2.27085 |
| 51091(IDI) | isopentenyl-diphosphate delta-isomerase | −1.77761 | −1.41504 | −2.58496 | −2 |
| 55836(DXS) | 1-deoxy-d-xylulose-5-phosphate synthase | −1.5009 | −3.98089 | −0.46063 | −3.64386 |
| 57427(DXR) | 1-deoxy-d-xylulose-5-phosphate reductoisomerase | −0.72763 | −2.5119 | −1.90923 | −1.48543 |
| 62474(CMK) | 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase | −1.08573 | −2.11548 | −0.74624 | −1.65605 |
| 39745(MDS) | 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase | 0.053771 | −2.21632 | −0.78022 | −1.05585 |
| 60959(HDS) | (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase | −0.8992 | −2.49304 | −0.91701 | −1.35311 |
| 7256(HDR) | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | −1.41504 | 2.357552 | −2 | −2 |
| 56508(HDR) | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | −0.94617 | −2.02185 | −0.83504 | −1.85798 |
| 4493(FPPS) | farnesyl diphosphate synthase | −0.11636 | −2.62272 | −1.76022 | −0.91222 |
| 21893(FPPS) | farnesyl diphosphate synthase | 0.584963 | −11.9658 | −11.9658 | −11.9658 |
| 5372GPPS | geranyl diphosphate synthase | 0.131245 | −0.93289 | −0.93289 | −1.80735 |
| 36997THS | Alpha-thujene synthase/sabinene synthase | −16.0102 | −0.23704 | −3.72247 | −0.83494 |
| 59387THS | Alpha-thujene synthase | −6.97871 | −0.70491 | −3.65678 | −0.9759 |
| 50359(GGPPS) | geranylgeranyl diphosphate synthase, type II | −0.42381 | −1.53343 | −1.6939 | −2.1964 |
| 52640(GGPPS) | geranylgeranyl diphosphate synthase, type II | 2 | 1.169925 | - | −0.41504 |
| 54221(GGPPS) | geranylgeranyl diphosphate synthase, type II | 0.211504 | 0.559427 | −0.24793 | 0.752072 |
| 57417(GGPPS) | geranylgeranyl diphosphate synthase, type II | −0.17834 | −4.77419 | −1.30288 | −2.77419 |
| 8148(GGPPS) | geranylgeranyl diphosphate synthase, type III | 13.77314 | - | - | - |
| 56393 | geranylgeranyl reductase | 0.688056 | 1.369234 | 2.863498 | 1.321928 |
| 58660 | geranylgeranyl reductase | 0.259087 | −1.28688 | −0.2303 | −1.84327 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, M.; Lin, L.; Chen, Y.; Wang, Y. Digital Gene Expression Profiling to Explore Differentially Expressed Genes Associated with Terpenoid Biosynthesis during Fruit Development in Litsea cubeba. Molecules 2016, 21, 1251. https://doi.org/10.3390/molecules21091251
Gao M, Lin L, Chen Y, Wang Y. Digital Gene Expression Profiling to Explore Differentially Expressed Genes Associated with Terpenoid Biosynthesis during Fruit Development in Litsea cubeba. Molecules. 2016; 21(9):1251. https://doi.org/10.3390/molecules21091251
Chicago/Turabian StyleGao, Ming, Liyuan Lin, Yicun Chen, and Yangdong Wang. 2016. "Digital Gene Expression Profiling to Explore Differentially Expressed Genes Associated with Terpenoid Biosynthesis during Fruit Development in Litsea cubeba" Molecules 21, no. 9: 1251. https://doi.org/10.3390/molecules21091251





