Identification of 3′,4′-Dimethoxy Flavonol-3-β-d-Glucopyranoside Metabolites in Rats by Liquid Chromatography-Electrospray Ionization Ion Trap Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of Chromatography and MS Conditions
2.2. Identification of DF3G
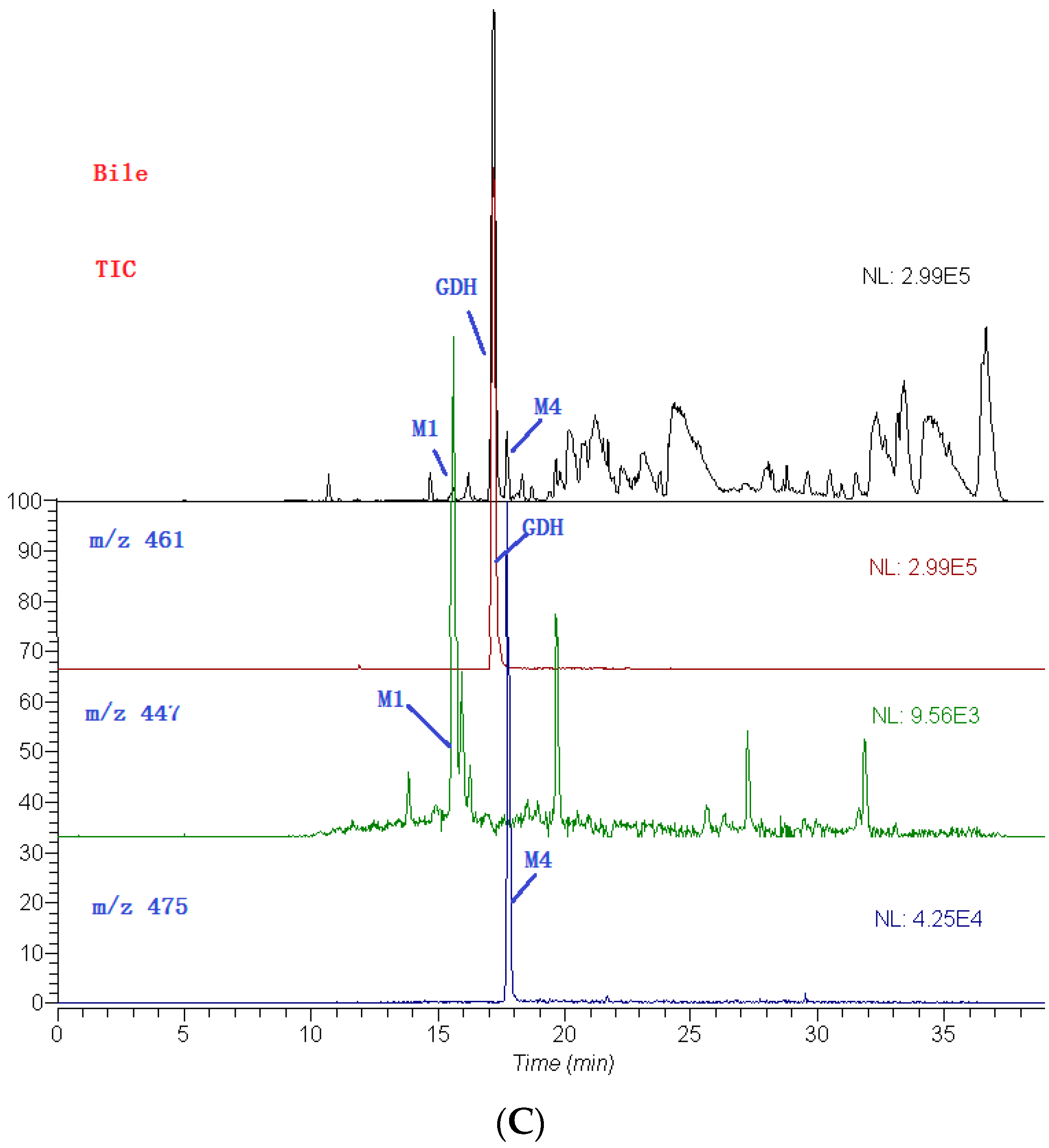
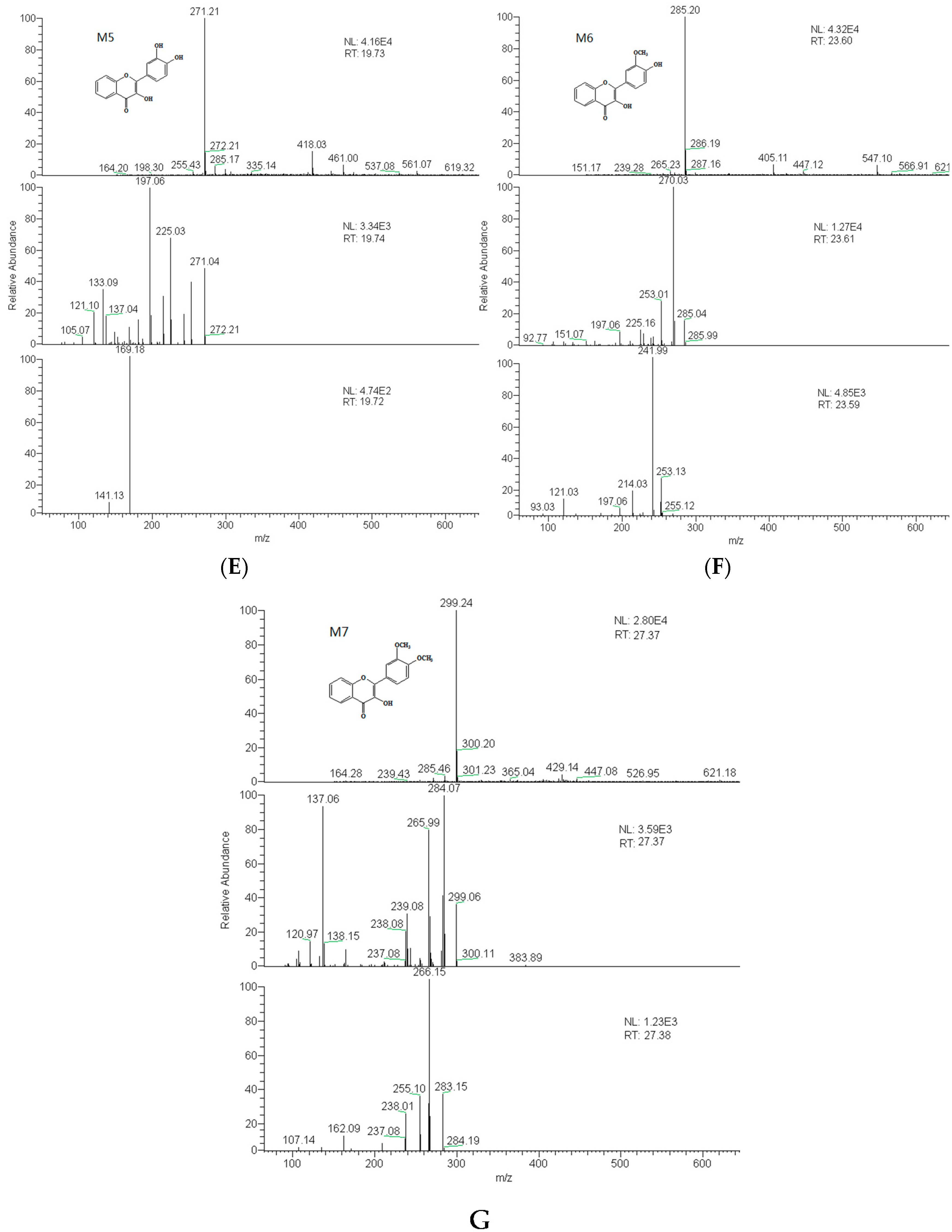
2.3. Identification of Metabolites
2.4. A Summary of Metabolic Pathways for DF3G
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Preparation of Standard Solutions
3.3. Animals and Drug Administration
3.4. Preparation of Urine, Feces and Bile Samples
3.5. Instrumentation
3.6. Liquid Chromatographic Conditions
3.7. Mass Spectrometer Conditions
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Morimoto, M.; Tanimoto, K.; Nakano, S.; Ozaki, T.; Nakano, A.; Komai, K. Insect Antifeedant Activity of Flavones and Chromones against Spodoptera litura. J. Agric. Food Chem. 2003, 51, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Galluzzo, P.; Ascenzi, P.; Bulzomi, P.; Marino, M. The nutritional flavanone naringenin triggers antiestrogenic effects by regulating estrogen receptor α-palmitoylation. Endocrinology 2008, 149, 2567–2575. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Wang, B.; Ren, P. Reduction of MTT by flavonoids in the absenceof cells. Colloids Surf. B 2005, 45, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Dong, B.; Yang, X.; Huang, C.; Wang, X.; Wu, X. Effect of lightonflavonoids biosynthesis in red rice Rdh. Agric. Sci. China 2009, 8, 746–752. [Google Scholar] [CrossRef]
- Procházková, D.; Boušová, I.; Wilhelmová, N. Antioxidant and prooxidantproperties of flavonoids. Fitoterapia 2011, 82, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Schijlen, E.G.; Ric, C.H.; Van, A.J.; Bovy, A.G. Modificationof flavonoid biosynthesis in crop plants. Phytochemistry 2004, 65, 2631–2648. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, C.H.; Moon, S.S.; Kim, E.; Kim, C.T.; Kim, B.H. Naringenin derivatives as anti-atherogenic agents. Bioorg. Med. Chem. Lett. 2003, 13, 3901–3903. [Google Scholar] [CrossRef] [PubMed]
- Romano, B.; Pagano, E.; Montanaro, V.; Fortunato, A.L.; Milic, N.; Borrelli, F. Novel Insights into the Pharmacology of Flavonoids. Phytother. Res. 2013, 27, 1588–1596. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.S.; Jiang, Y.Y.; Wu, Q.Y.; Liu, G.Q.; Dai, Y.; Liu, W.B. 3-Substituted Oxo-3,4-Two Methoxy Flavonoids with Reducing Blood Lipid. China Patent ZL 200810034446.0, 20 August 2008. [Google Scholar]
- Chan, S.A.; Chen, M.J.; Liu, T.Y.; Fu, M.R.; Deng, J.F. Determination of aristolochic acids in medicinal plant and herbal product by liquid chromatography-electrospray-ion trap mass spectrometry. Talanta 2003, 60, 679–685. [Google Scholar] [CrossRef]
- Yu, X.; Cui, D.; Davis, M.R. Identification of in vitro metabolites of Indinavir by “intelligent automated LC-MS/MS” (INTAMS) utilizing triple quadrupole tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 1999, 10, 175–183. [Google Scholar] [CrossRef]
- Lam, W.R.; Ramanathan, R. In electrospray ionization source hydrogen/deuterium exchange LC-MS and LC-MS/MS for characterization of metabolites. J. Am. Soc. Mass Spectrom. 2002, 13, 345–353. [Google Scholar] [CrossRef]
- Gu, J.K.; Zhong, D.F.; Chen, X.Y. Analysis of O-glucuronide conjugates in urine by electrospray ion trap mass spectrometry. Fresenius J. Anal. Chem. 1999, 365, 553–558. [Google Scholar] [CrossRef]
- Day, A.J.; Cañada, F.J.; Díaz, J.C.; Kroon, P.A.; Mclauchlan, R.; Faulds, C.B.; Plumb, G.W.; Morgan, M.R.; Williamson, G. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. FEBS Lett. 2000, 468, 166–170. [Google Scholar] [CrossRef]
- Van, W.H.; Boersma, M.G.; Vervoort, J.; Rietjens, I.M. Identification of 14 quercetin phase II mono- and mixed conjugates and their formation by rat and human phase II in vitro model systems. Chem. Res. Toxicol. 2004, 17, 1520–1530. [Google Scholar]
- Gradolatto, A.; Canyvenc-Lavier, M.C.; Basly, J.P.; Siess, M.H.; Teyssier, C. Metabolism of apigenin by liver phase I and phase II enzymes and by isolated perfused rat liver. Drug Metab. Dispos. 2004, 32, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhang, Z.C.; Fu, G.; Sun, S.F. Liquid chromatography-tandem mass spectrometry analysis of protocatechuic aldehyde and its phase I and II metabolites in rat. J. Chromatogr. B 2007, 856, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, F.; Isabelle, R. Determination of Flavone, Flavonol, and Flavanone Aglycones by Negative Ion Liquid Chromatography Electrospray Ion Trap Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar]
- Xu, Y.; Dong, J.; Wand, H.; Wan, L.R.; Duan, Y.S.; Chen, S.Z. Fragmentation study of the C ring in flavone and isoflavone aglycones by electrospray ion trap time-of-flight mass spectrometry. Chem. J. Chin. Univ. 2009, 30, 46–50. [Google Scholar]
- Bhagwat, P.; Amit, G.; Hardik, T.; Saranjit, S. Metabolite identification by liquid chromatography-mass spectrometry. Trends. Anal. Chem. 2011, 30, 360–387. [Google Scholar]
- Zheng, G.S.; Zhao, X.; Fan, G.R. Study on the mass fragmentation pathway of flavonol glucose glycoside and its structural analogues. Pharm. Care Res. 2015, 15, 196–200. [Google Scholar]
- Breinholt, V.M.; Oford, E.A.; Brouwer, C.; Nielsen, S.E.; Brøsen, K.; Friedberg, T. In vitro investigation of cytochrome P450-mediated metabolism of dietary flavonoids. Food Chem Toxicol. 2002, 40, 609–616. [Google Scholar] [CrossRef]
- Jin, Y.; Wu, L.; Tang, Y.P.; Cao, Y.J.; Li, S.J.; Shen, J.; Yue, S.; Qu, C.; Shan, C.; Cui, X.; et al. UFLC-Q-TOF/MS based screening and identification of the metabolites in plasma, bile, urine and feces of normal and blood stasis rats after oral administration of hydroxysafflor yellow A. J. Chromatogr. B 2016, 1012, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.P.; Gee, J.M.; Dupont, M.S.; Needs, P.W.; Mellon, F.A.; Williamson, G.; Johnson, I.T. Hydrolysis by lactase phlorizin hydrolase is the first step in the uptake of daidzein glucosides by rat small intestine in vitro. Xenobiotica 2003, 33, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Sesink, A.L.; Arts, I.C.; Faassen-Peters, M.; Hollman, P.C. Intestinal uptake of quercetin-3-glucoside in rats involves hydrolysis by lactase phlorizin hydrolase. J. Nutr. 2003, 133, 773–776. [Google Scholar] [PubMed]
- Nemeth, K.; Plumb, G.W.; Berrin, J.G.; Juge, N.; Jacob, R.; Naim, H.Y.; Williamson, G.; Swallow, D.M.; Kroon, P.A. Deglycosylation by small intestinal epithelial cell beta-glucosidases is a critical step in the absorption and metabolism of dietary flavonoid glycosides in humans. Eur. J. Nutr. 2003, 42, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cui, L.; Duan, X.; Ma, B.; Zhong, D. Pharmacokinetics and metabolism of the flavonoid scutellarin in humans after a single oral administration. Drug Metab. Dispos. 2006, 34, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the DF3G are available from the authors.
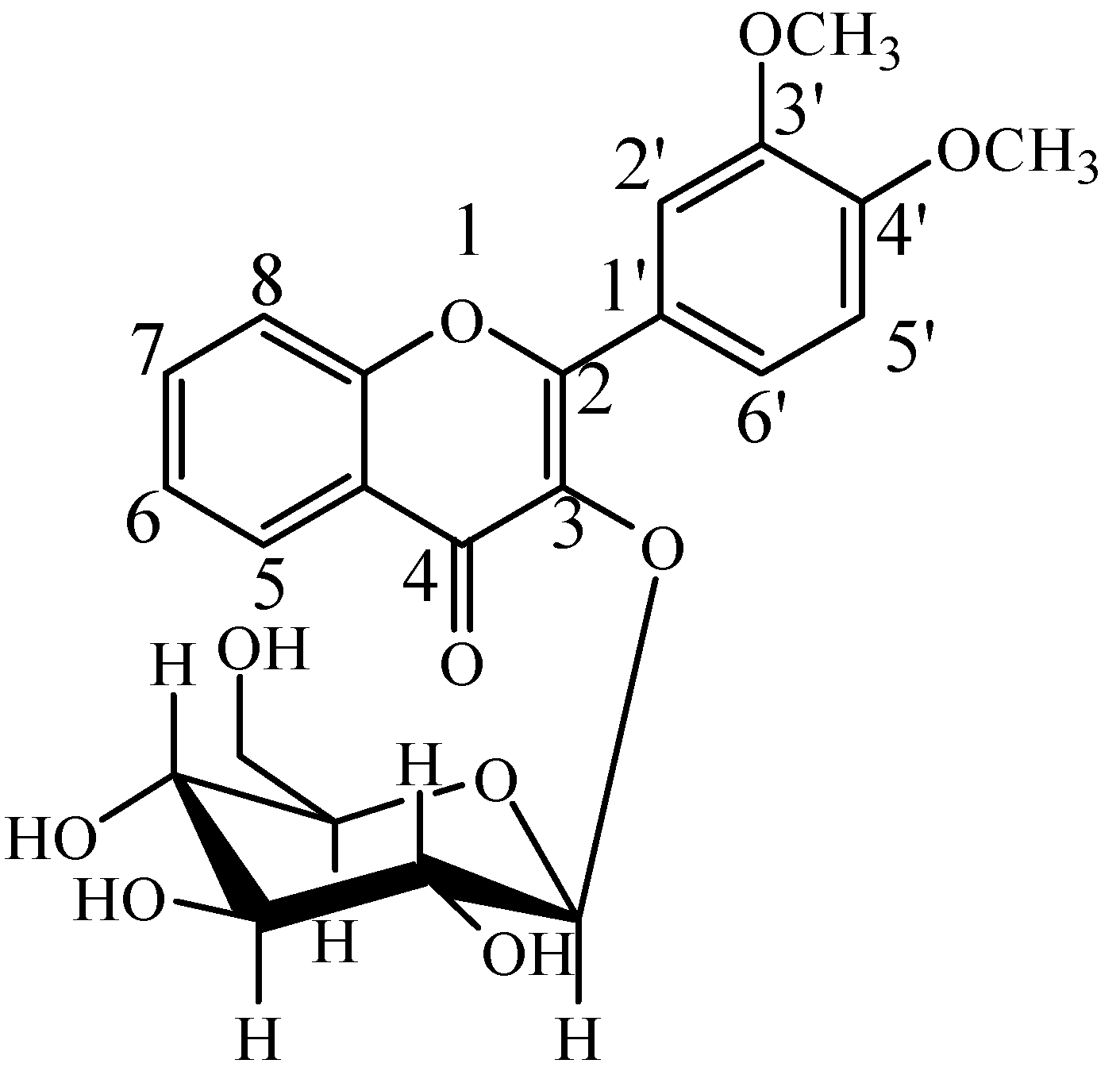
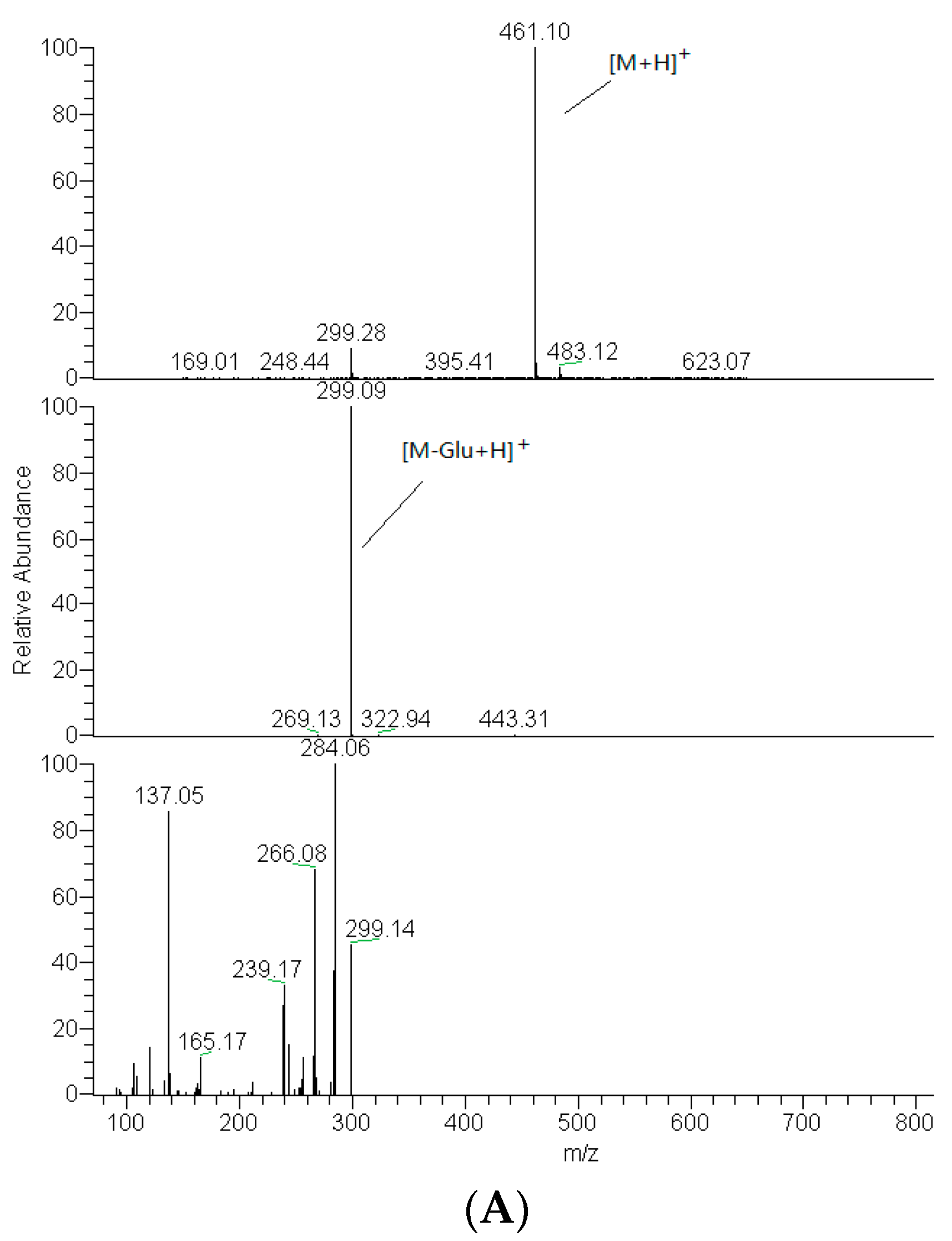



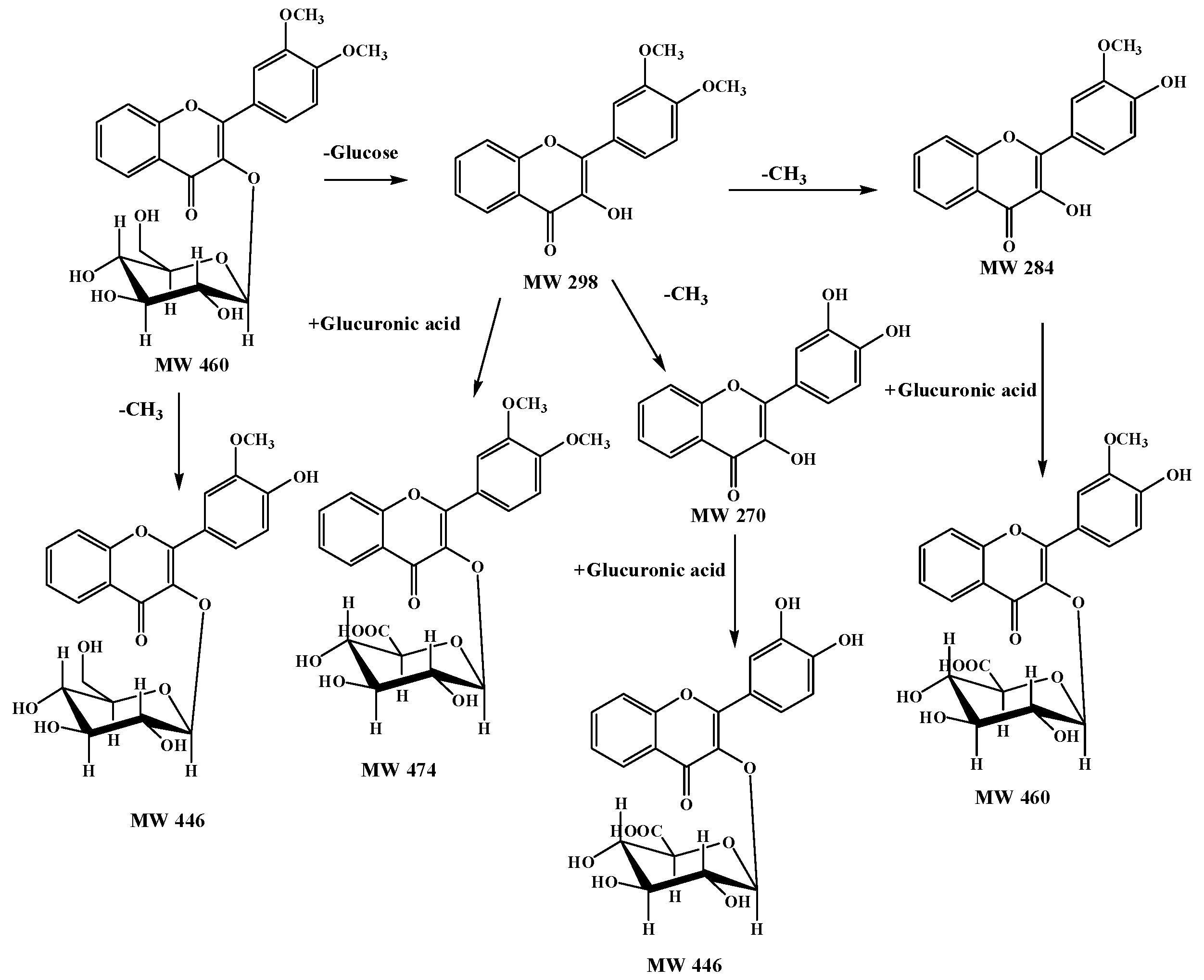
| No. | tR (min) | Compound Name | MW | Formula | Parent Ions (m/z) | MSn (m/z) |
|---|---|---|---|---|---|---|
| M1 | 15.6 | 3′-methoxy-4′-hydroxy flavonol-3-O-β-d-glucopyranoside | 446 | C22H22O10 | 447 | 285, 270 |
| M2 | 15.9 | 3′,4′-dihydroxyflavonol-3-O-β-d-glucuronide | 446 | C22H22O10 | 447 | 271, 225, 197 |
| M3 | 16.2 | 3′-methoxy-4′-hydroxy flavonol-3-O-β-d-glucopyranoside | 460 | C23H24O10 | 461 | 285, 270 |
| M4 | 17.8 | 3′,4′-dimethoxy flavonol-3-O-β-d-glucopyranoside | 474 | C23H22O11 | 475 | 299, 284, 266 |
| M5 | 19.7 | 3′,4′-dihydroxy flavonol | 270 | C15H10O5 | 271 | 225, 197, 169, 270, 242 |
| M6 | 23.6 | 3′-methoxy-4′-hydroxy flavonol | 284 | C16H12O5 | 285 | 270, 242 |
| M7 | 27.4 | 3′,4′-dimethoxy flavonol | 298 | C17H14O5 | 299 | 284, 266 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wen, J.; Cao, Y.; Jiang, Y.; Huang, J.; Fan, G.; Lou, Y. Identification of 3′,4′-Dimethoxy Flavonol-3-β-d-Glucopyranoside Metabolites in Rats by Liquid Chromatography-Electrospray Ionization Ion Trap Mass Spectrometry. Molecules 2016, 21, 470. https://doi.org/10.3390/molecules21040470
Zhu Y, Wen J, Cao Y, Jiang Y, Huang J, Fan G, Lou Y. Identification of 3′,4′-Dimethoxy Flavonol-3-β-d-Glucopyranoside Metabolites in Rats by Liquid Chromatography-Electrospray Ionization Ion Trap Mass Spectrometry. Molecules. 2016; 21(4):470. https://doi.org/10.3390/molecules21040470
Chicago/Turabian StyleZhu, Yuan, Jun Wen, Yuqing Cao, Yuanying Jiang, Jinghua Huang, Guorong Fan, and Yuefen Lou. 2016. "Identification of 3′,4′-Dimethoxy Flavonol-3-β-d-Glucopyranoside Metabolites in Rats by Liquid Chromatography-Electrospray Ionization Ion Trap Mass Spectrometry" Molecules 21, no. 4: 470. https://doi.org/10.3390/molecules21040470





