The Role of CD44 and ERM Proteins in Expression and Functionality of P-glycoprotein in Breast Cancer Cells
Abstract
:1. Introduction
2. Results
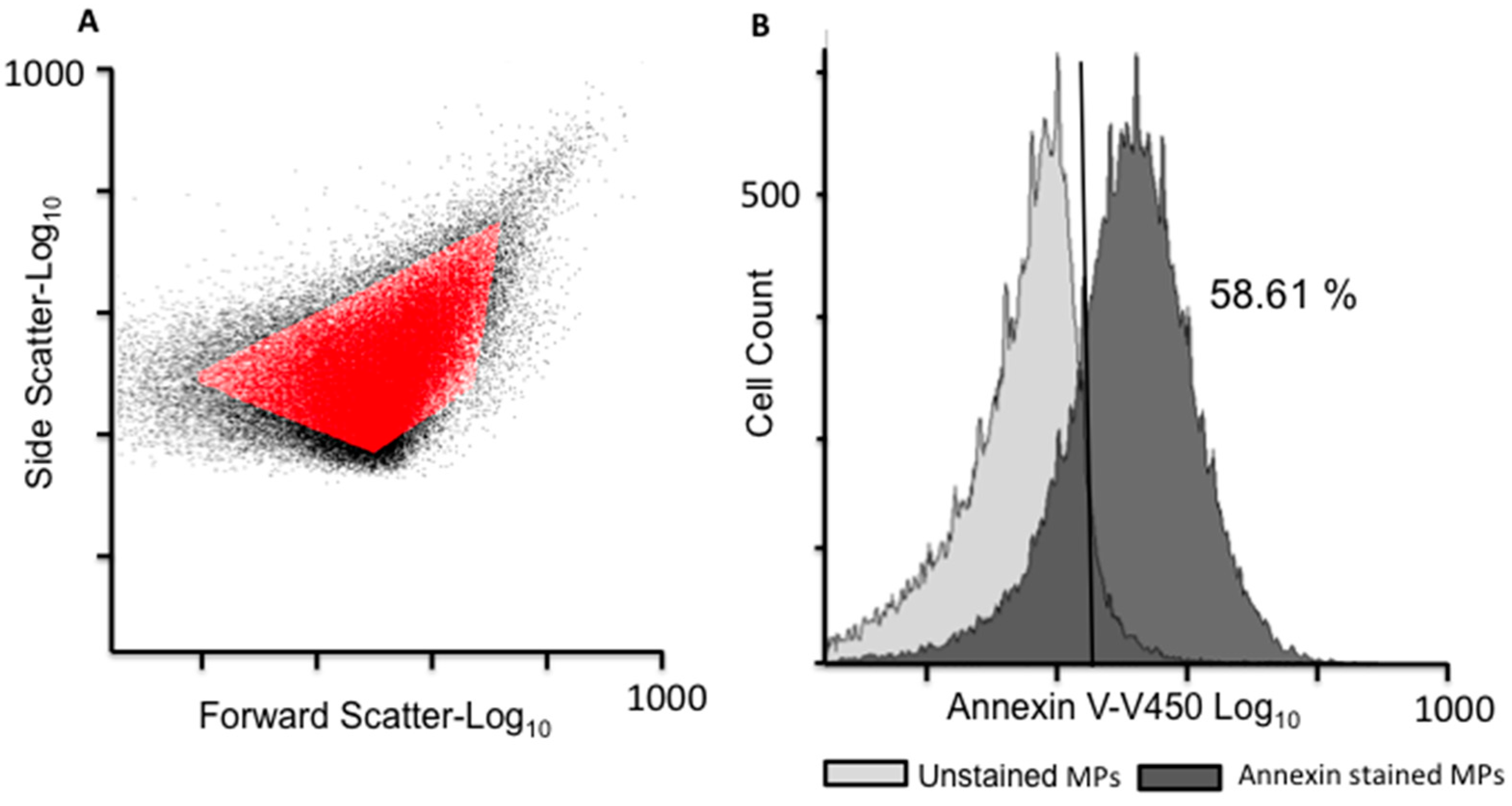
2.1. EV Isolation and Validation
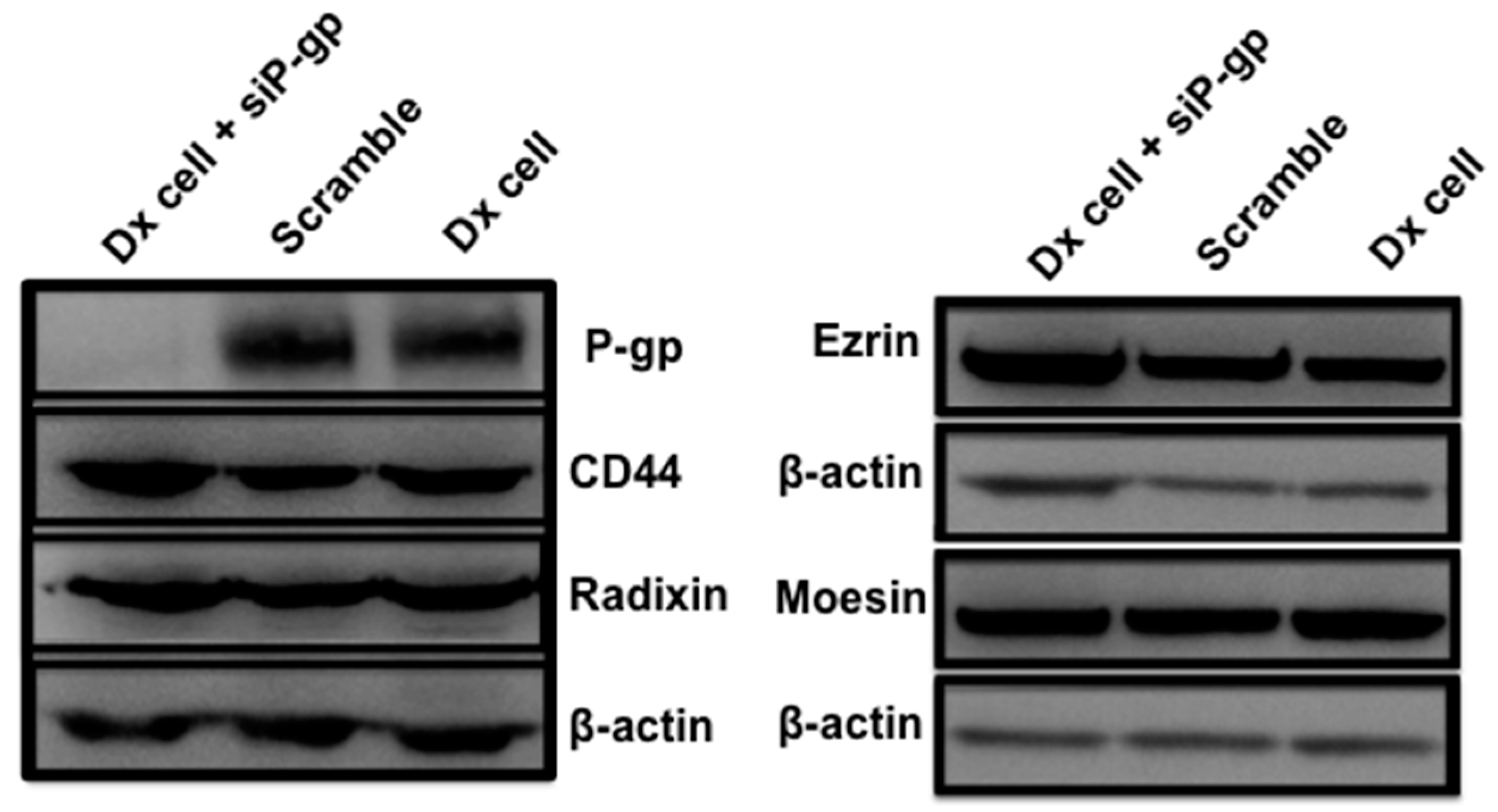
2.2. Proteomic Profiling by LC/MS/MS and Western Blotting
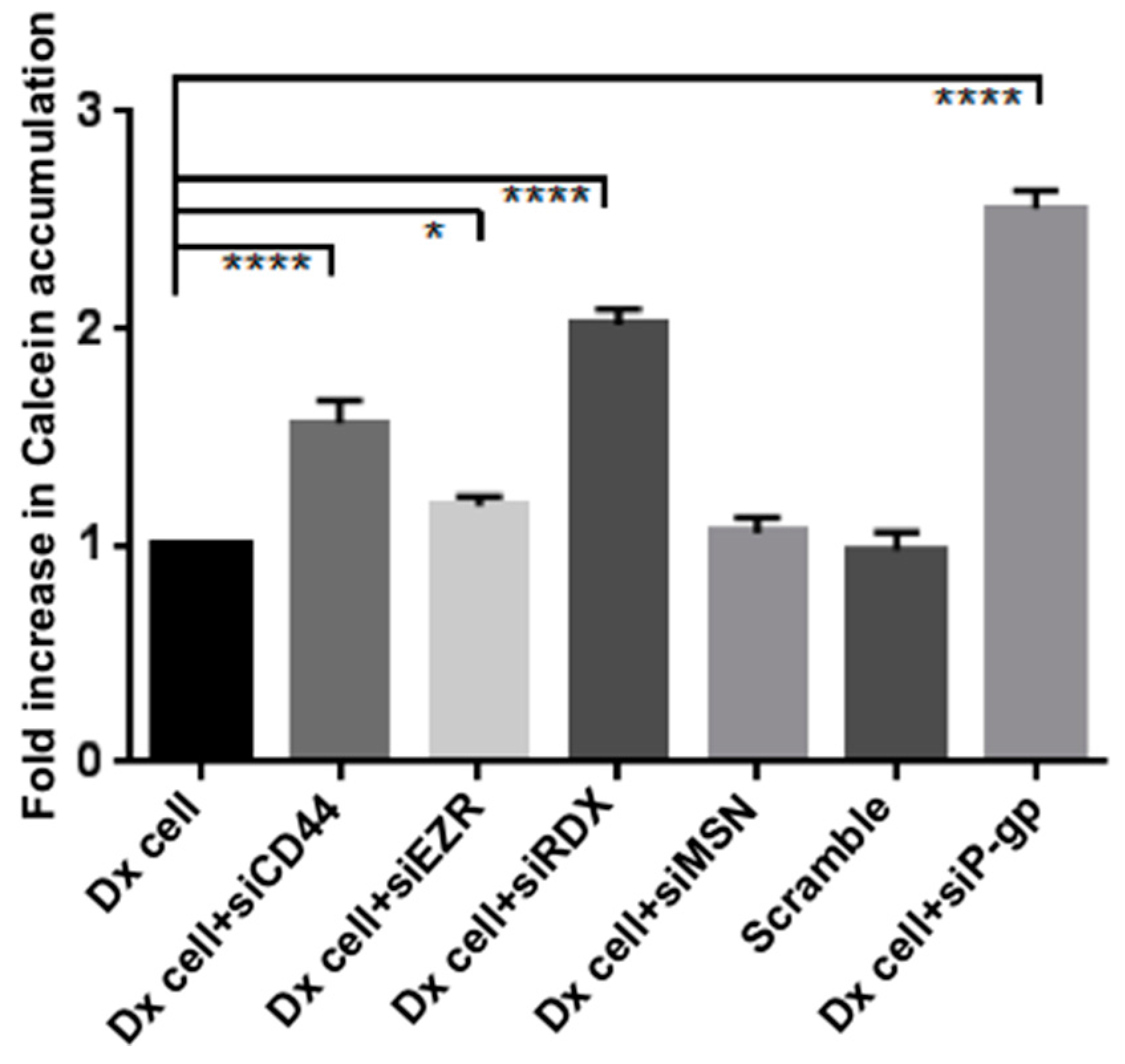
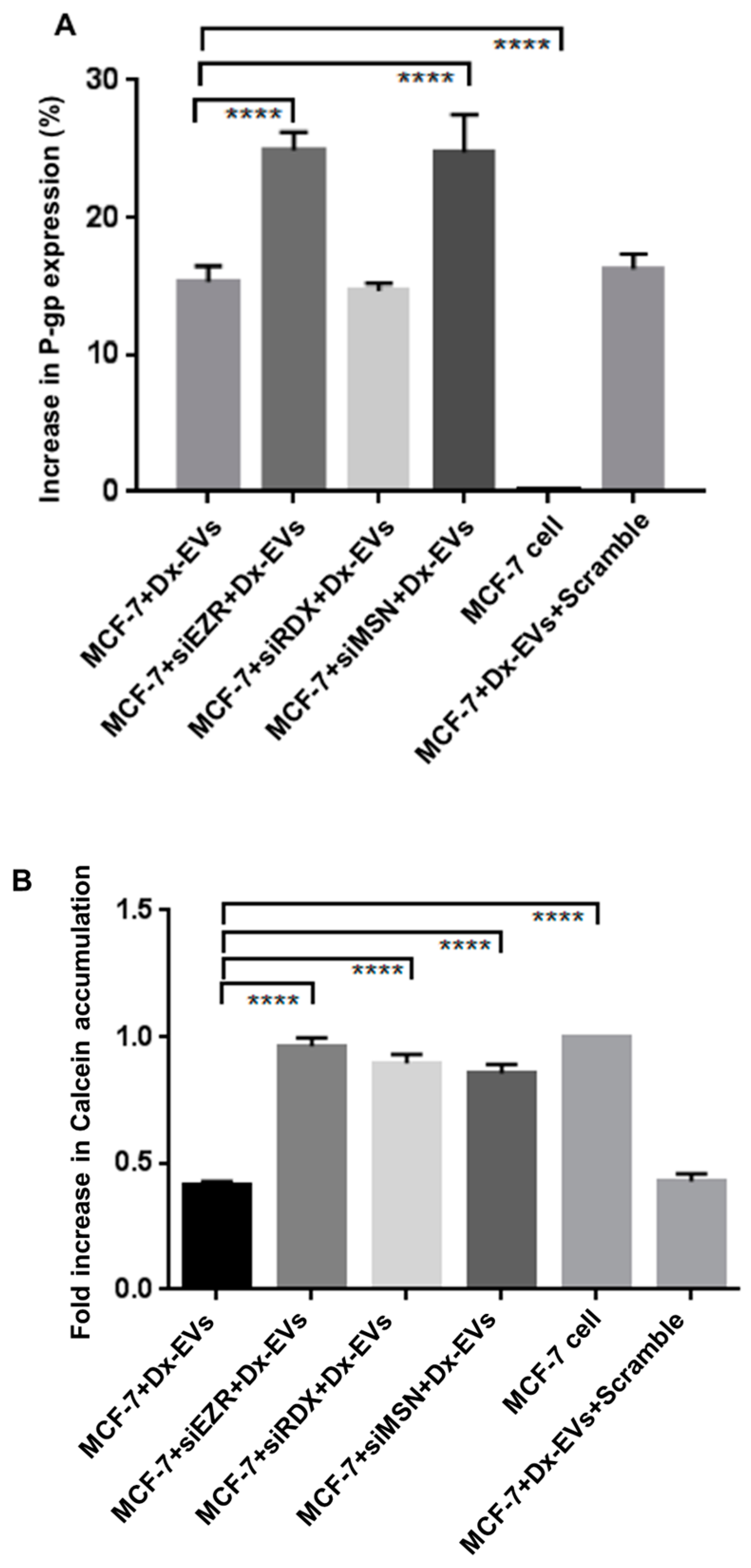
2.3. Gene Silencing of ERM and CD44 Affects P-gp Drug Efflux in Resistant Breast Cancer Cells
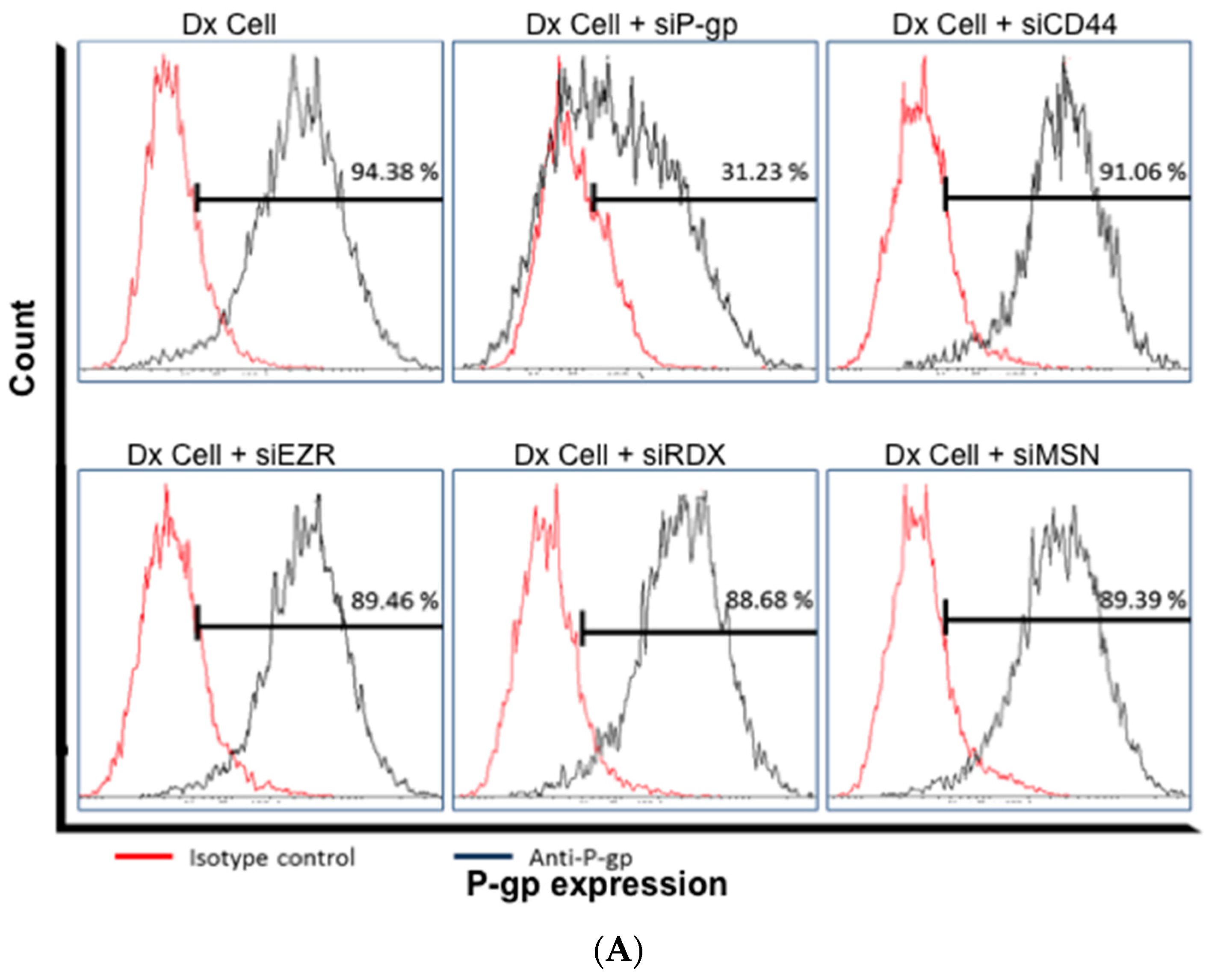
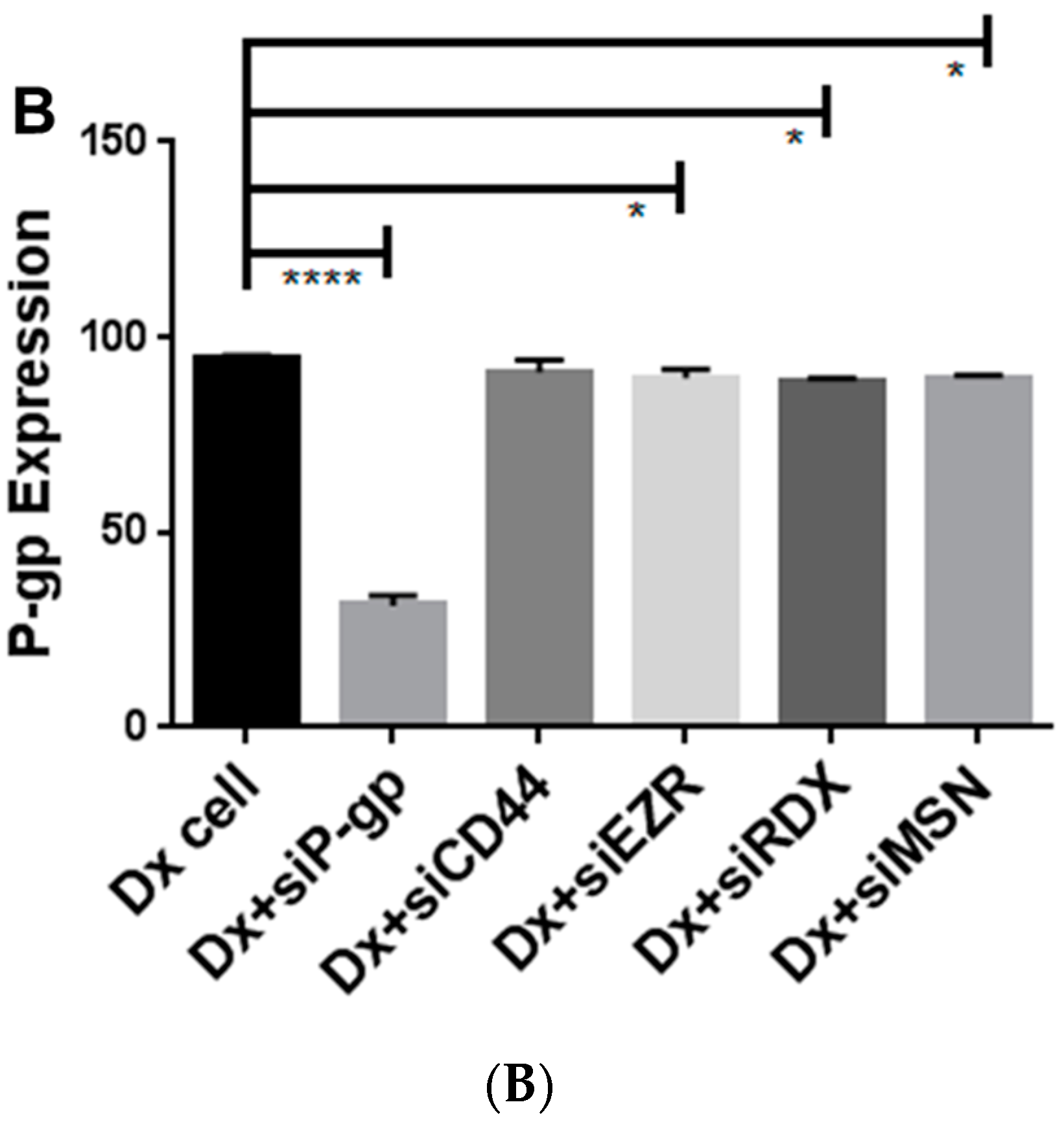
2.4. Gene Silencing of P-gp Has no Effect on ERM and CD44 Expression in Resistant Breast Cancer Cells
2.5. Role of ERM in the Vesicular Transfer of P-gp to Recipient Drug Sensitive Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Harvesting EVs
4.3. Protein Extraction
4.4. SDS-PAGE and Trypsin In-Gel Digestion
4.5. LC/MS/MS
4.6. Data Analysis
4.7. In Vitro Silencing
4.8. Calcein-AM Dye Exclusion Assay
4.9. Western Blotting
4.10. Vesicular Transfer Experiments
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ERM | Ezrin-Radixin-Moesin |
| EVs | Extracellular vesicles |
| MDR | Multidrug resistance |
| P-gp | P-glycoprotein |
References
- Pokharel, D.; Padula, M.P.; Lu, J.F.; Tacchi, J.L.; Luk, F.; Djordjevic, S.P.; Bebawy, M. Proteome analysis of multidrug-resistant, breast cancer-derived microparticles. J. Extracell. Vesicles 2014. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.F.; Pokharel, D.; Bebawy, M. Mrp1 and its role in anticancer drug resistance. Drug Metabol. Rev. 2015, 47, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Bebawy, M.; Combes, V.; Lee, E.; Jaiswal, R.; Gong, J.; Bonhoure, A.; Grau, G. Membrane microparticles mediate transfer of p-glycoprotein to drug sensitive cancer cells. Leukemia 2009, 23, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.F.; Luk, F.; Gong, J.; Jaiswal, R.; Grau, G.E.; Bebawy, M. Microparticles mediate mrp1 intercellular transfer and the re-templating of intrinsic resistance pathways. Pharmacol. Res. 2013, 76, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Luk, F.; Jaiswal, R.; George, A.M.; Grau, G.E.R.; Bebawy, M. Microparticle drug sequestration provides a parallel pathway in the acquisition of cancer drug resistance. Eur. J. Pharmacol. 2013, 721, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Luk, F.; Jaiswal, R.; Bebawy, M. Microparticles mediate the intercellular regulation of microrna-503 and proline-rich tyrosine kinase 2 to alter the migration and invasion capacity of breast cancer cells. Front. Oncol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.; Gong, J.; Sambasivam, S.; Combes, V.; Mathys, J.-M.; Davey, R.; Grau, G.E.; Bebawy, M. Microparticle-associated nucleic acids mediate trait dominance in cancer. FASEB J. 2012, 26, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Bebawy, M.; Morris, M.; Roufogalis, B. Selective modulation of p-glycoprotein-mediated drug resistance. Br. J. Cancer 2001, 85. [Google Scholar] [CrossRef] [PubMed]
- Bebawy, M.; Morris, M.B.; Roufogalis, B.D. A continuous fluorescence assay for the study of p-glycoprotein-mediated drug efflux using inside-out membrane vesicles. Anal. Biochem. 1999, 268, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Jaiswal, R.; Dalla, P.; Luk, F.; Bebawy, M. Microparticles in cancer: A review of recent developments and the potential for clinical application. Semin. Cell Dev. Biol. 2015, 40, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Luciani, F.; Molinari, A.; Lozupone, F.; Calcabrini, A.; Lugini, L.; Stringaro, A.; Puddu, P.; Arancia, G.; Cianfriglia, M.; Fais, S. P-glycoprotein–actin association through erm family proteins: A role in p-glycoprotein function in human cells of lymphoid origin. Blood 2002, 99, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Puddu, P.; Fais, S.; Luciani, F.; Gherardi, G.; Dupuis, M.L.; Romagnoli, G.; Ramoni, C.; Cianfriglia, M.; Gessani, S. Interferon-gamma up-regulates expression and activity of p-glycoprotein in human peripheral blood monocyte-derived macrophages. Lab. Investig. J. Tech. Methods Pathol. 1999, 79, 1299–1309. [Google Scholar]
- Fu, D.; Roufogalis, B.D. Actin disruption inhibits endosomal traffic of p-glycoprotein-egfp and resistance to daunorubicin accumulation. Am. J. Physiol. Cell Physiol. 2007, 292, C1543–C1552. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, D.; Zamboni, S.; Federici, C.; Lugini, L.; Lozupone, F.; Milito, A.D.; Cecchetti, S.; Cianfriglia, M.; Fais, S. P-glycoprotein binds to ezrin at amino acid residues 149–242 in the ferm domain and plays a key role in the multidrug resistance of human osteosarcoma. Int. J. Cancer 2012, 130, 2824–2834. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.; Luk, F.; Dalla, P.V.; Grau, G.E.R.; Bebawy, M. Breast cancer-derived microparticles display tissue selectivity in the transfer of resistance proteins to cells. PLoS ONE 2013, 8, e61515. [Google Scholar] [CrossRef] [PubMed]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Miletti-González, K.E.; Chen, S.; Muthukumaran, N.; Saglimbeni, G.N.; Wu, X.; Yang, J.; Apolito, K.; Shih, W.J.; Hait, W.N.; Rodríguez-Rodríguez, L. The cd44 receptor interacts with p-glycoprotein to promote cell migration and invasion in cancer. Cancer Res. 2005, 65, 6660–6667. [Google Scholar] [CrossRef] [PubMed]
- Yonemura, S.; Hirao, M.; Doi, Y.; Takahashi, N.; Kondo, T.; Tsukita, S.; Tsukita, S. Ezrin/radixin/moesin (erm) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of cd44, cd43, and icam-2. J. Cell Biol. 1998, 140, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Oishi, K.; Sato, N.; Sagara, J.; Kawai, A. Erm family members as molecular linkers between the cell surface glycoprotein cd44 and actin-based cytoskeletons. J. Cell Biol. 1994, 126, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M. String 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Roseblade, A.; Luk, F.; Ung, A.; Bebawy, M. Targeting microparticle biogenesis: A novel approach to the circumvention of cancer multidrug resistance. Curr. Cancer Drug Targets 2015, 15, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. Panther: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Aneja, R.; Chandra, R.; Lage, H.; Joshi, H.C. Reversal of p-glycoprotein–mediated multidrug resistance in cancer cells by the c-jun nh2-terminal kinase. Cancer Res. 2006, 66, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Goebeler, M.; Kaufmann, D.; Brocker, E.; Klein, C.E. Migration of highly aggressive melanoma cells on hyaluronic acid is associated with functional changes, increased turnover and shedding of cd44 receptors. J. Cell Sci. 1996, 109, 1957–1964. [Google Scholar] [PubMed]
- Ambudkar, S.V.; Kimchi-Sarfaty, C.; Sauna, Z.E.; Gottesman, M.M. P-glycoprotein: From genomics to mechanism. Oncogene 2003, 22, 7468–7485. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Coley, H.M. Overcoming multidrug resistance in cancer: An update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control 2003, 10, 159–165. [Google Scholar] [PubMed]
- Kano, T.; Wada, S.; Morimoto, K.; Kato, Y.; Ogihara, T. Effect of knockdown of ezrin, radixin, and moesin on p-glycoprotein function in hepg2 cells. J. Pharm. Sci. 2011, 100, 5308–5314. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Boulan, E.; Nelson, W.J. Morphogenesis of the polarized epithelial cell phenotype. Science 1989, 245, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Moyer, B.D.; Duhaime, M.; Shaw, C.; Denton, J.; Reynolds, D.; Karlson, K.H.; Pfeiffer, J.; Wang, S.; Mickle, J.E.; Milewski, M. The pdz-interacting domain of cystic fibrosis transmembrane conductance regulator is required for functional expression in the apical plasma membrane. J. Biol. Chem. 2000, 275, 27069–27074. [Google Scholar] [PubMed]
- Toole, B.P. Hyaluronan-cd44 interactions in cancer: Paradoxes and possibilities. Clin. Cancer Res. 2009, 15, 7462–7468. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.; Peyrollier, K.; Xia, W.; Gilad, E. Hyaluronan-cd44 interaction activates stem cell marker nanog, stat-3-mediated mdr1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J. Biol. Chem. 2008, 283, 17635–17651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiao, R.; Xiong, J.; Leng, J.; Ehtisham, A.; Hu, Y.; Ding, Q.; Xu, H.; Liu, S.; Wang, J. Activated erm protein plays a critical role in drug resistance of molt4 cells induced by ccl25. PLoS ONE 2013, 8, e52384. [Google Scholar] [CrossRef] [PubMed]
- Turunen, O.; Wahlström, T.; Vaheri, A. Ezrin has a cooh-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol. 1994, 126, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Cain, J.W.; Hauptschein, R.S.; Stewart, J.K.; Bagci, T.; Sahagian, G.G.; Jay, D.G. Identification of cd44 as a surface biomarker for drug resistance by surface proteome signature technology. Mol. Cancer Res. 2011, 9, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Toole, B.P.; Slomiany, M.G. Hyaluronan, cd44 and emmprin: Partners in cancer cell chemoresistance. Drug Resist. Updates 2008, 11, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Bebawy, M.; Kable, E.P.; Roufogalis, B.D. Dynamic and intracellular trafficking of p-glycoprotein-egfp fusion protein: Implications in multidrug resistance in cancer. Int. J. Cancer 2004, 109, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Bogema, D.R.; Deutscher, A.T.; Woolley, L.K.; Seymour, L.M.; Raymond, B.B.; Tacchi, J.L.; Padula, M.P.; Dixon, N.E.; Minion, F.C.; Jenkins, C. Characterization of cleavage events in the multifunctional cilium adhesin mhp684 (p146) reveals a mechanism by which mycoplasma hyopneumoniae regulates surface topography. mBio 2012, 3, e00282-11. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, T.; Ohga, N.; Akiyama, K.; Hirata, N.; Kitahara, S.; Maishi, N.; Osawa, T.; Yamamoto, K.; Kondoh, M.; Shindoh, M.; et al. Tumor-derived microvesicles induce proangiogenic phenotype in endothelial cells via endocytosis. PLoS ONE 2012, 7, e34045. [Google Scholar]
- Perkins, D.; Pappin, D.; Creasy, D.; Cottrell, J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by ms/ms and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Sample Availability: Not available.

© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokharel, D.; Padula, M.P.; Lu, J.F.; Jaiswal, R.; Djordjevic, S.P.; Bebawy, M. The Role of CD44 and ERM Proteins in Expression and Functionality of P-glycoprotein in Breast Cancer Cells. Molecules 2016, 21, 290. https://doi.org/10.3390/molecules21030290
Pokharel D, Padula MP, Lu JF, Jaiswal R, Djordjevic SP, Bebawy M. The Role of CD44 and ERM Proteins in Expression and Functionality of P-glycoprotein in Breast Cancer Cells. Molecules. 2016; 21(3):290. https://doi.org/10.3390/molecules21030290
Chicago/Turabian StylePokharel, Deep, Matthew P. Padula, Jamie F. Lu, Ritu Jaiswal, Steven P. Djordjevic, and Mary Bebawy. 2016. "The Role of CD44 and ERM Proteins in Expression and Functionality of P-glycoprotein in Breast Cancer Cells" Molecules 21, no. 3: 290. https://doi.org/10.3390/molecules21030290






