Improving Free Radical Scavenging Activity of Soy Isoflavone Glycosides Daidzin and Genistin by 3′-Hydroxylation Using Recombinant Escherichia coli
Abstract
:1. Introduction
2. Results
2.1. Construction of Recombinant E. coli Expressing Tyrosinase from B. megaterium
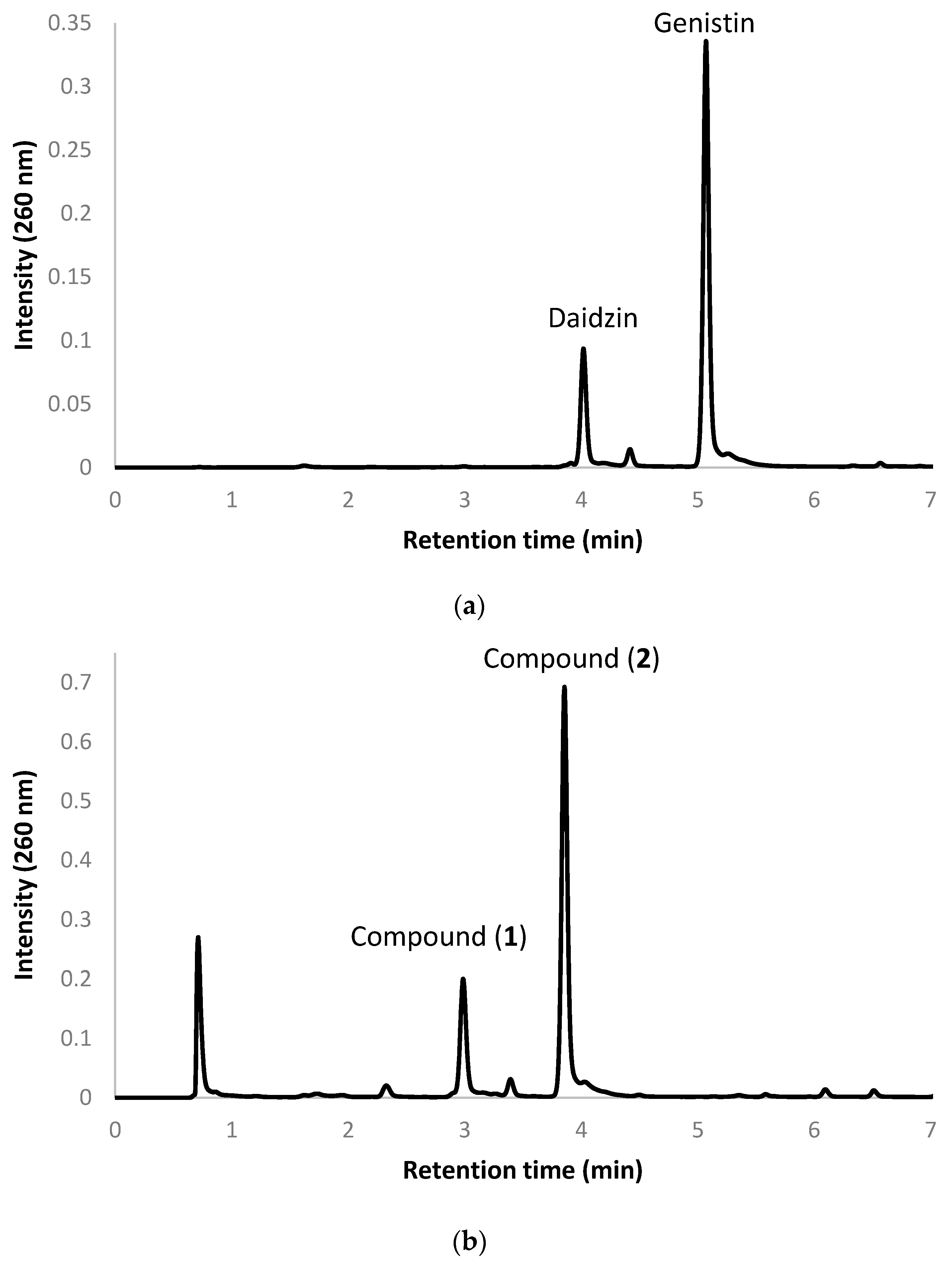
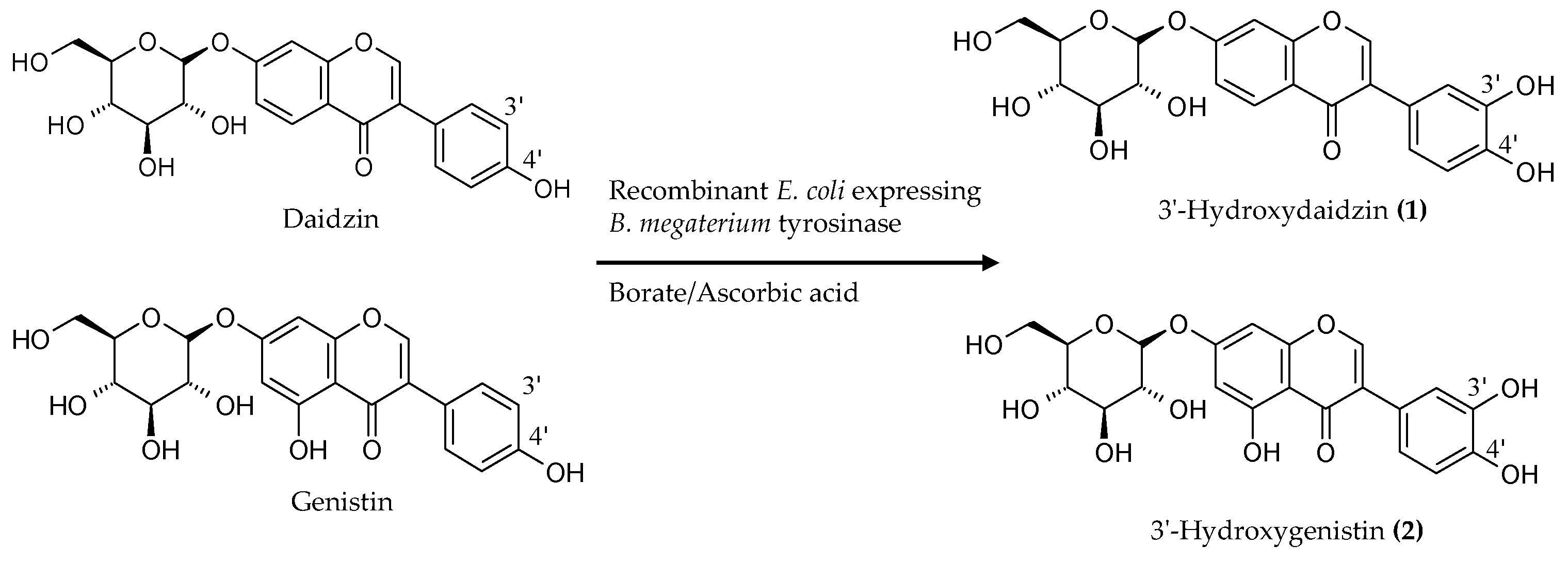
2.2. Biotransformation of a Soy Isoflavone Crude Extract by the Recombinant E. coli and Purification and Identification of the Biotransformation Products
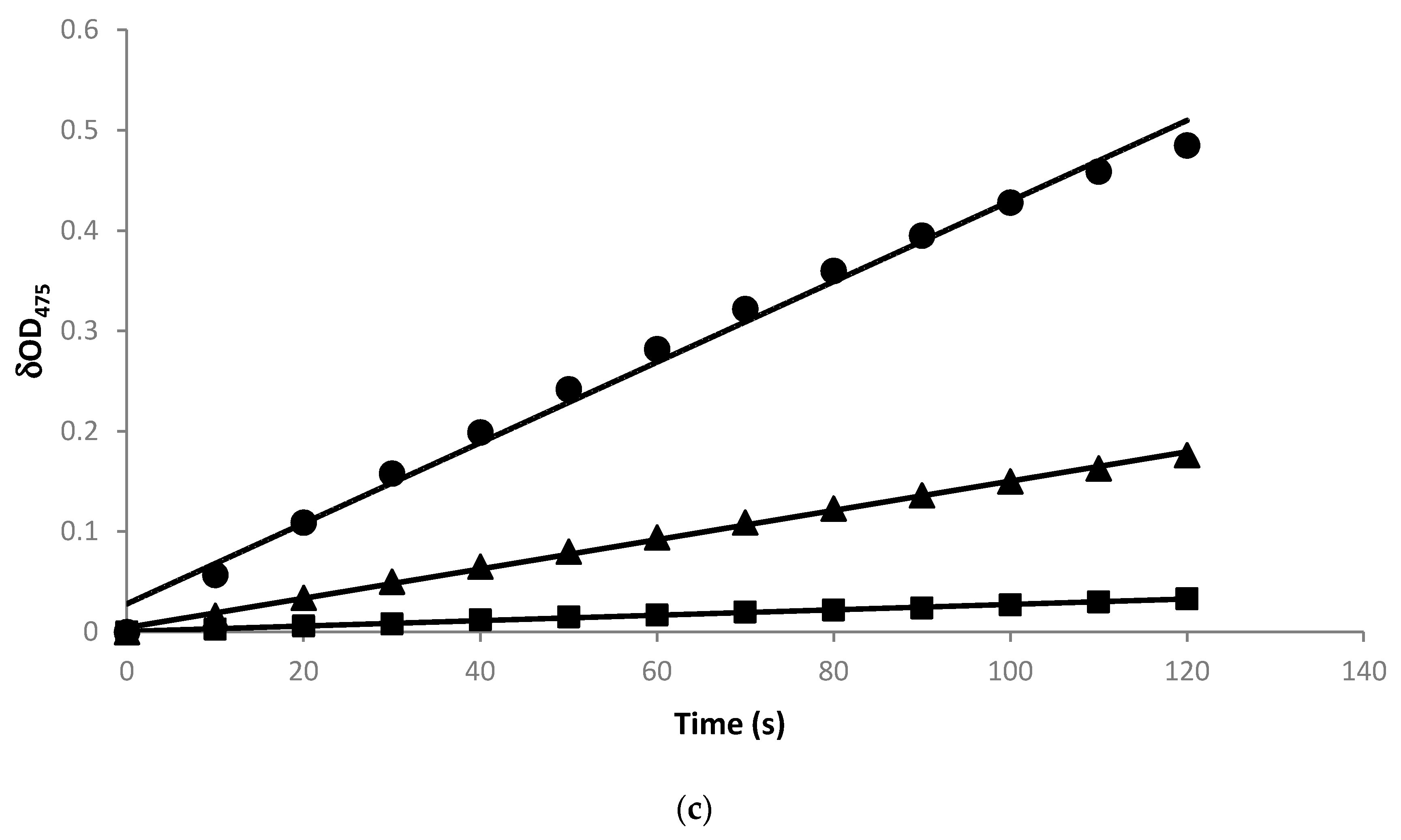
2.3. Radical Scavenging Activity of the Biotransformation Products
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Chemicals
4.2. Expression of B. megaterium Tyrosinase in E. coli
4.3. Tyrosinase Activity Assay
4.4. Biotransformation and UPLC Analysis
4.5. Purification and Identification of Biotransformation Products
4.6. DPPH Free Radical Scavenging Activity Assay
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Franke, A.A.; Custer, L.J.; Cerna, C.M.; Narala, K.K. Quantitation of phytoestrogens in legumes by HPLC. J. Agric. Food Chem. 1994, 42, 1905–1913. [Google Scholar] [CrossRef]
- Vitale, D.C.; Piazza, C.; Melilli, B.; Drago, F.; Salomone, S. Isoflavones: estrogenic activity, biological effect and bioavailability. Eur. J. Drug Metab. Pharmacokinet. 2013, 38, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-S. Isolation, bioactivity, and production of ortho-hydroxydaidzein and ortho-hydroxygenistein. Int. J. Mol. Sci. 2014, 15, 5699–5716. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S.; Chao, S.Y.; Chen, Y.C. Production of ortho-hydroxydaidzein derivatives by a recombinant strain of Pichia pastoris harboring a cytochrome P450 fusion gene. Process Biochem. 2013, 48, 426–429. [Google Scholar] [CrossRef]
- Chiang, C.-M.; Ding, H.-Y.; Lu, J.-Y.; Chang, T.-S. Biotransformation of isoflavones daidzein and genistein by recombinant Pichia pastoris expressing membrane-anchoring and reductase fusion chimeric CYP105D7. J. Taiwan Inst. Chem. Eng. 2016, 60, 26–31. [Google Scholar] [CrossRef]
- Lee, H.; Kim, B.G.; Ahn, J.H. Production of bioactive hydroxyflavones by using monooxygenase from Saccharothrix espanaensis. J. Biotechnol. 2014, 176, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Baek, K.; Lee, J.E.; Kim, B.G. Using tyrosinase as a monophenol monooxygenase: A combined strategy for effective inhibition of melanin formation. Biotechnol. Bioeng. 2016, 113, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, I.; Jedrejek, D.; Ciesla, L.; Pecio, L.; Masullo, M.; Piacente, S.; Oleszek, W.; Stochmal, A. Isolation, chemical and free radical scavenging characterization of phenolics from Trifolium scabrum L. aerial parts. J. Agric. Food Chem. 2013, 61, 4417–4423. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Bae, E.Y.; Song, J.H.; Baek, S.H.; Kwon, D.H. Inhibitory effects of orobol 7-O-d-glucoside from banaba (Lagerstroemia speciosa L.) on human rhinoviruses replication. Lett. Appl. Microbiol. 2010, 51, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Y.; Tsai, Y.-H.; Yu, I.-Z.; Chang, T.-S. Improving 3′-hydroxygenistein production in recombinant Pichia pastoris by a periodic hydrogen-shocking strategy. J. Microbiol. Biotechnol. 2016, 26, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S. An updated review of tyrosinase inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S.; Ding, H.Y.; Tai, S.S.K.; Wu, C.Y. Tyrosinase inhibitors isolated from soygerm koji fermented with Aspergillus oryzae BCRC 32288. Food Chem. 2007, 105, 1430–1438. [Google Scholar] [CrossRef]
- Choi, K.Y.; Park, H.Y.; Kim, B.G. Characterization of bi-functional CYP154 from Nocardia farcinica IFM10152 in the O-deakylation and ortho-hydroxylation of formononetin. Enzym. Microb. Technol. 2010, 47, 327–334. [Google Scholar] [CrossRef]
- Cao, G.H.; Sofic, E.; Prior, R.L. Antioxidant and prooxidant behavior of flavonoids: structure-activity relationships. Free Radic. Biol. Med. 1997, 22, 749–760. [Google Scholar] [CrossRef]
- Ye, H.; Xu, H.; Yu, C.; Dai, Y.; Liu, G.; Xu, W.; Yuan, S. Hydroxylation of naringin by Trichoderma harzianum to dramatically improve its antioxidative activity. Enzym. Microb. Technol. 2009, 45, 282–287. [Google Scholar] [CrossRef]
- Liu, G.; Sun, L.; Wang, S.; Chen, C.; Guo, T.; Ji, Y.; Li, X.; Huang, G.; Wei, H.; Dai, Y.; Yuan, S. Hydroxylation modification and free radical scavenging activity of puerarin-7-O-fructoside. Folia Microb. 2011, 56, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Fussell, K.C.; Udasin, R.G.; Smith, P.J.S.; Gallo, M.A.; Laskin, J.D. Catechol metabolites of endogeneous estrogens induce redox cycling and generate reactive oxygen species in breast epithelial cells. Carcinogenesis 2011, 32, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds 3′-hydroxydaidzin and 3′-hydroxygenistin are available from the authors.


© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, C.-M.; Wang, D.-S.; Chang, T.-S. Improving Free Radical Scavenging Activity of Soy Isoflavone Glycosides Daidzin and Genistin by 3′-Hydroxylation Using Recombinant Escherichia coli. Molecules 2016, 21, 1723. https://doi.org/10.3390/molecules21121723
Chiang C-M, Wang D-S, Chang T-S. Improving Free Radical Scavenging Activity of Soy Isoflavone Glycosides Daidzin and Genistin by 3′-Hydroxylation Using Recombinant Escherichia coli. Molecules. 2016; 21(12):1723. https://doi.org/10.3390/molecules21121723
Chicago/Turabian StyleChiang, Chien-Min, Dong-Sheng Wang, and Te-Sheng Chang. 2016. "Improving Free Radical Scavenging Activity of Soy Isoflavone Glycosides Daidzin and Genistin by 3′-Hydroxylation Using Recombinant Escherichia coli" Molecules 21, no. 12: 1723. https://doi.org/10.3390/molecules21121723




