Damxungmacin A and B, Two New Amicoumacins with Rare Heterocyclic Cores Isolated from Bacillus subtilis XZ-7
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation of the Compounds
2.2. Evaluation of Anti-Antibacterial Activities
2.3. Anti-Proliferative Assay on Human Tumor Cells
3. Experimental Section
3.1. General Information
3.2. Bacterial Material
3.3. Fermentation, Extraction and Isolation
3.4. Spectroscopic Data of Compounds
3.5. Cytotoxicity Assay In Vitro
3.6. Antibacterial Assay In Vitro
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| UV | Ultraviolet |
| IR | Infrared |
| NMR | Nuclear magnetic resonance |
| HR-ESI-MS | High resolution electrospray ionization mass spectroscopy |
| RP-MPLC | Reverse-phase medium-oressure liquid chromatography |
| UPLC-DAD-MS | Ultra-performance liquid chromatography coupled with diode array detection and mass spectroscopy |
References
- Han, X.Y.; Liu, S.W.; Wang, F.F.; Liu, J.M.; Hu, X.X.; You, X.F.; Shang, G.D.; Zhang, Y.B.; Sun, C.H. Research Progress of Amicoumacin Group Antibiotic. World Notes Antibiot. 2013, 34, 106–115. [Google Scholar]
- Irina, V.P.; Philippe, B.; Bernard, V.; Bernard, F.; Irina, B.S.; Francis, M.; Maria, C.U. In vitro anti-helicobacter pylori Activity of the Probiotic Strain Bacillus subtilis 3 Is Due to Secretion of Antibiotics. Antimicrob. Agents Chemother. 2001, 45, 3156–3161. [Google Scholar]
- Jiro, I.; Takashi, S.; Shoji, O.; Shinji, M.; Yasukatsu, Y.; Uichi, S.; Shigeharu, I. Isolation, physicochemical properties and biological activities of amicoumacins produced by Bacillus pumilus. Agric. Biol. Chem. 1982, 46, 1255–1259. [Google Scholar]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.Y. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Librada, C.H.; Cristina, A.S.; Dolores, G.G. Antitumor Isocoumarins. U.S. Patent 5,925,671, 20 July 1999. [Google Scholar]
- Librada, M.C.; Jose, L.F.P.; Julia, P.B. PM-94128, a new isocoumarin antitumor agent produced by a marine bacterium. J. Antibiot. 1997, 50, 175–176. [Google Scholar]
- Tutomu, S.; Koji, N.; Kenichi, S.; Moto, M.; Takeshi, S. A new isocoumarin antibiotic, Y-05460M-A. J. Antibiot. 1992, 45, 1949–1952. [Google Scholar]
- Jiro, I.; Shoji, O.; Nobusuke, N.; Yoshio, K.; Shigeharu, I. Chemical structures of amicoumacins produced by Bacillus pumilus. Agric. Biol. Chem. 1982, 46, 2659–2665. [Google Scholar]
- Yukiji, S.; Hiroshi, H. 1H-2-Benzopyran-1-one derivatives, microbial products with pharmacological activity. Relationship between structure and activity in 6-[1(S)-(3(S),4-Dihydro-8-hydroxy-l-oxo-1H-2-benzopyran-3-yl)-3-methylbutyl]-amino]-4(S),5(S)-aih ydroxy-6-oxo-3(S)-ammoniohexanoate. J. Med. Chem. 1983, 26, 1370–1374. [Google Scholar]
- Yukiji, S.; Hiroshi, H.; Tadaaki, O.; Mitsuru, S. Structures and The Chemical Nature of AI-77s. Tetrahedron Lett. 1982, 23, 5435–5438. [Google Scholar]
- Yukiji, S.; Hiroshi, H.; Tadaaki, O.; Mitsuru, S. Studies on AI-77s, microbial products with gastroprotective activity. Tetrahedron 1984, 40, 2519–2527. [Google Scholar]
- Miwa, A.; Ken-ichi, O.; Tsuyoshi, F.; Michinori, T.; Ryuji, Y.; Tamotsu, F.; Yasuhiro, I. Bacilosarcins A and B, novel bioactive isocoumarins with unusual heterocyclic cores from the marine-derived bacterium Bacillus subtilis. Tetrahedron 2008, 64, 6420–6425. [Google Scholar]
- Liu, S.; Han, X.; Jiang, Z.; Wu, G.; Hu, X.; You, X.; Jiang, J.; Zhang, Y.; Sun, C. Hetiamacin B–D, new members of amicoumacin group antibiotics isolated from Bacillus subtilis PJS. J. Antibiot. 2016, 69, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.W.; Jin, J.; Chen, C.; Liu, J.M.; Li, J.Y.; Wang, F.F.; Jiang, Z.K.; Hu, J.H.; Gao, Z.X.; Yao, F.; et al. PJS, a novel isocoumarin with hexahydropyrimidine ring from Bacillus subtilis PJS. J. Antibiot. 2013, 66, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Liu, S.W.; Zhou, Z.H.; Tuo, L.; Ling, B.; Guo, L.; Dong, Y.P.; Wang, F.F.; Jiang, Z.K.; Li, X.J.; et al. Studies on amicoumacin group antibiotics produced by Bacillus strain XZ7. Chin. J. Antibiot. 2014, 39, 801–807. [Google Scholar]
- Liu, S.W.; Xia, M.; Gao, Q.; Jiang, Z.K.; Hu, X.X.; He, Q.Y.; You, X.F.; Zhang, Y.Q.; Sun, C.H. Isolation, identification and bioactivity of amicoumacin group antibiotics produced by Bacillus strain XZ7 isolated from Tibet. Nat. Prod. Res. Dev. 2016, 28, 5–10. [Google Scholar]
- Pantelis, C.; Vassiliki, G.K.; Constantinos, G.T.; Andreas, G.T.; Michael, S.; Ioannis, P.G. 1H-NMR as a structural and analytical tool of intra- and inter-molecular hydrogen bonds of phenol containing natural products and model compounds. Molecules 2014, 19, 13643–13682. [Google Scholar]
- Vassiliki, G.K.; Panteli, C.; Alexandra, P.; Charalambos, G.P.; Vassiliki, E.; Andreas, G.T.; Ioannis, P.G. Hydrogen bonding probes of phenol –OH groups. Org. Biomol. Chem. 2013, 11, 1013–1025. [Google Scholar]
- Daniela, R.; Eva, L.; Alexander, O.B.; Helge, B.B. A new type of pyrrolidine biosynthesis is involved in the late steps of xenocoumacin production in Xenorhabdus nematophila. ChemBioChem. 2009, 10, 1997–2001. [Google Scholar]
- Dongjin, P.; Kristin, C.; Ransome, H.; Swati, S.; Daniela, R.; Helge, B.B.; Steven, F. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol. Microbiol. 2009, 73, 938–949. [Google Scholar]
- Yong, X.L.; Zhong, R.L.; Kazuya, Y.; Ying, X.; Wei, P.Z.; Hera, V.; Roberto, K.; Bradley, S.M.; Pei, Y.Q. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci. Rep. 2015, 5, 9383. [Google Scholar] [CrossRef]
- Tu, Y.Y.; Ni, M.Y.; Zhong, Y.R.; Li, L.; Gui, S.L.; Zhang, M.Q.; WANG, X.Z.; Liang, X.T. Studies on the constituents of artemisia annul L. Acta Pharm. Sin. 1981, 16, 366–370. [Google Scholar]
- Chen, Q.; Zhu, L.H.; Xu, Y.F.; Fan, J.Z. A new steroidal alkaloid from the bulbus of Fritillaria wabuensia. Acta Pharm. Sin. 2004, 39, 348–350. [Google Scholar]
- Yan, J.; Liu, Z.Q.; Zhu, H.G.; Liu, S.Y. Analysis of two trace alkaloid isomers in strychnos nux-vomical by electrospray ionization tandem mass spectrometry. Acta Chim. Sin. 2007, 65, 49–52. [Google Scholar]
- Clinical and Laboratory Standards Institute. M7-M9: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Dai, Y.H.; Shen, B.; Xia, M.Y.; Wang, A.D.; Chen, Y.L.; Liu, D.C.; Wang, D. A new indole alkaloid from the toad venom of bufo bufo gargarizans. Molecules 2016, 21, 349. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Sample of the compound Damxungmacin A is available from the authors, but sample of the compound Damxungmacin B is not available.
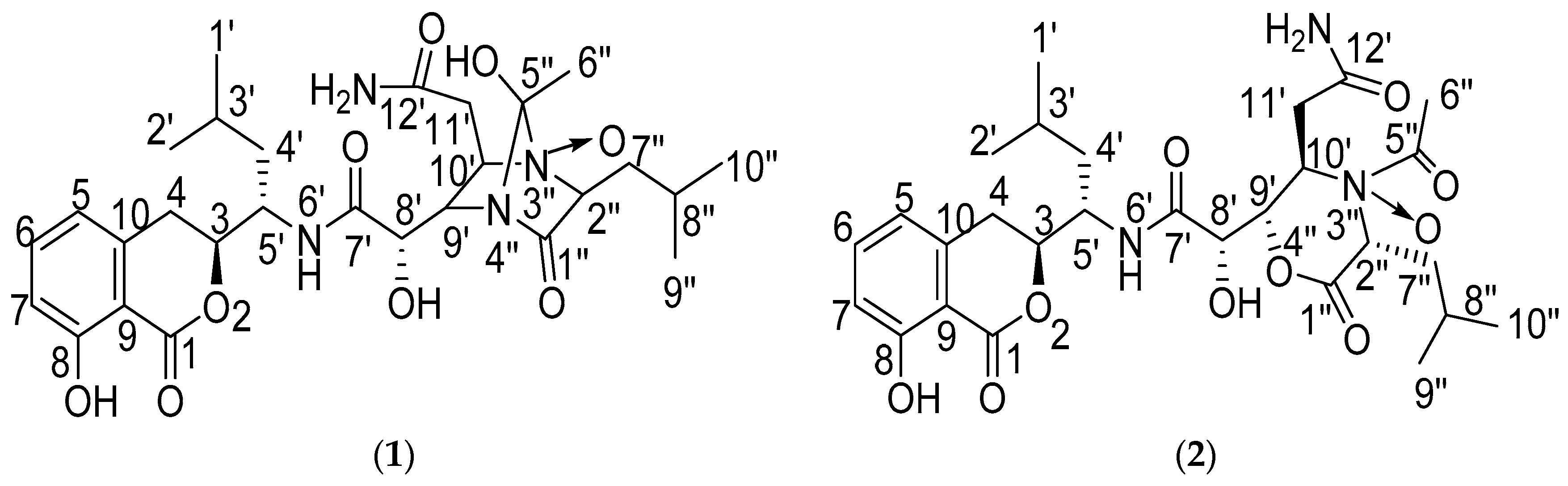
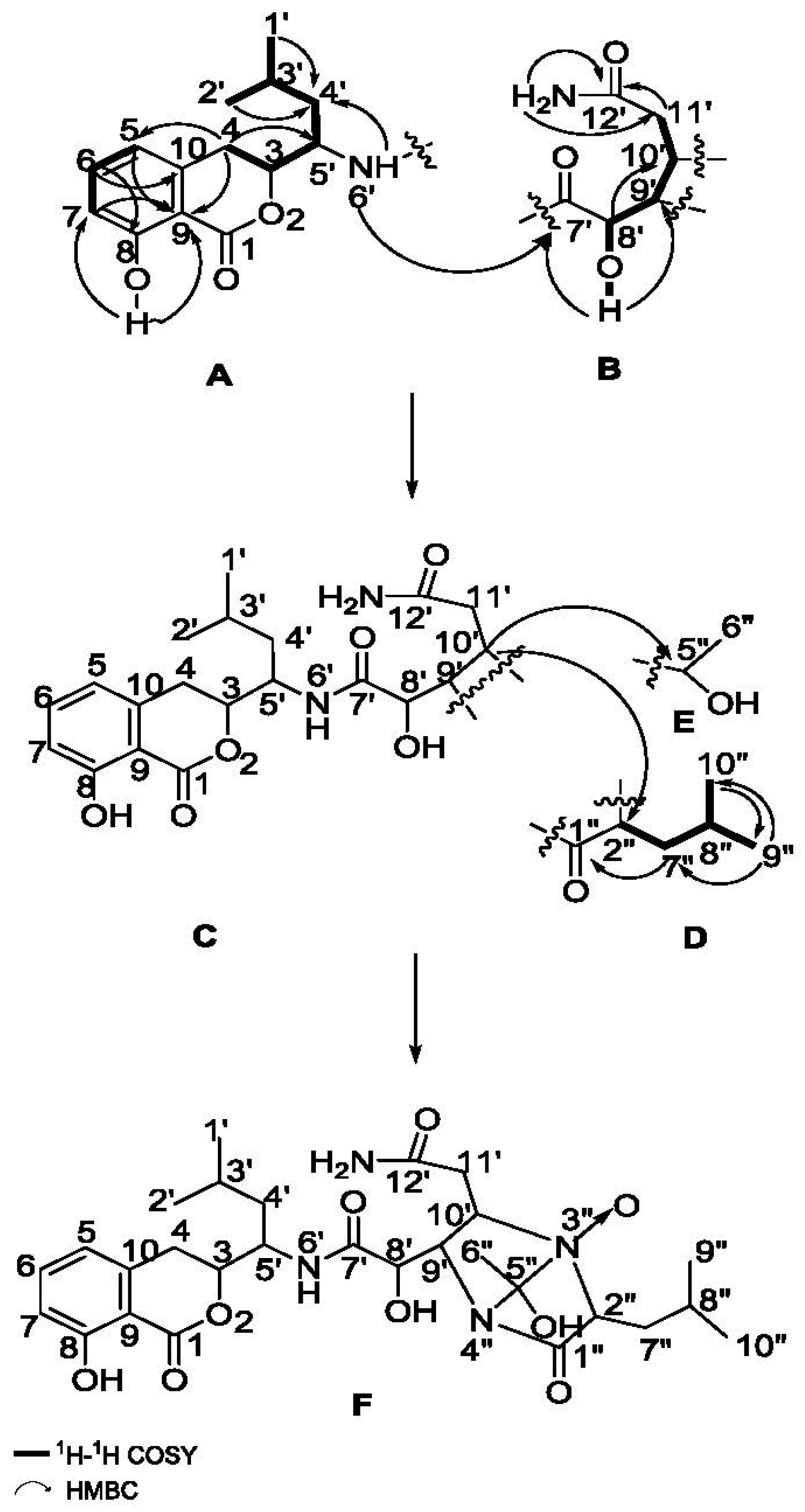
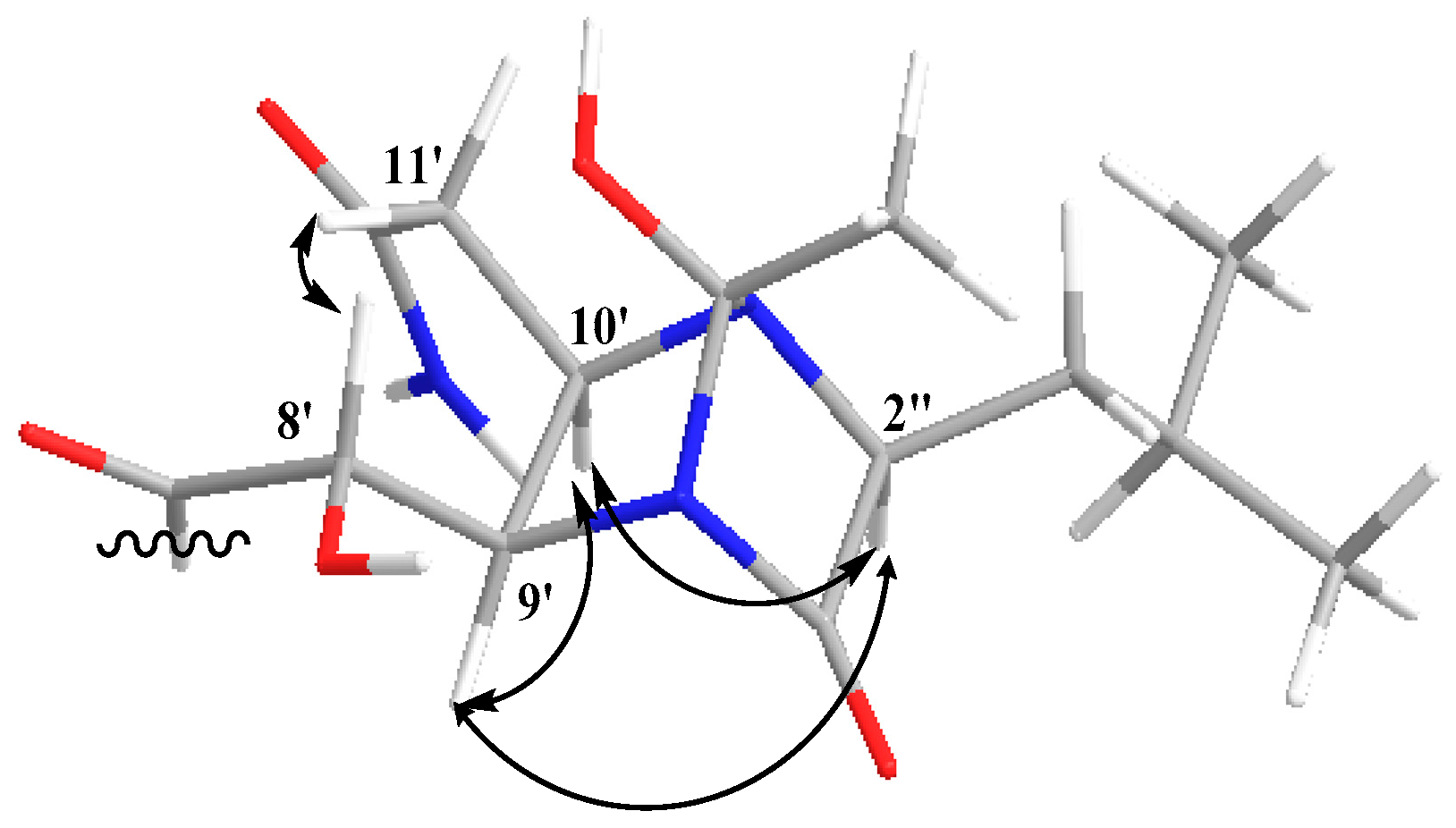


| No. | Damxungmacin A (1) | Damxungmacin B (2) | ||||
|---|---|---|---|---|---|---|
| δC a | δH (mult, J in Hz) b | HMBC | δC a | δH (mult, J in Hz) b | HMBC | |
| 1 | 169.4 | 168.9 | ||||
| 3 | 81.4 | 4.69 (1H, d, J = 12.6 Hz) | 10 | 80.6 | 4.68 (1H, d, J = 8.4 Hz) | |
| 4 | 29.0 | 3.22 (1H, t, J = 12.6 Hz) 2.77 (1H, d, J = 16.8 Hz) | 3,5,9,10,5′ | 29.1 | 2.99 (1H, t, J = 12.6 Hz) 2.90 (1H, d, J = 12.6 Hz) | 3,9,10 |
| 5 | 118.7 | 6.79 (1H, d, J = 7.2 Hz) | 4,7,8,9 | 118.4 | 6.82 (2H, unresolved multiplet) | |
| 6 | 136.2 | 7.45 (1H, t, J = 7.2 Hz) | 7,8,9,10 | 136.3 | 7.48 (1H, unresolved multiplet) | |
| 7 | 114.9 | 6.81 (1H, d, J = 7.2 Hz) | 1,5,6,8,9,10 | 115.4 | 6.84 (2H, unresolved multiplet) | |
| 8 | 160.7 | 161.0 | ||||
| 8-OH | 10.82 (1H, s) | 7,8,9 | Unidentified | |||
| 9 | 108.2 | 108.4 | ||||
| 10 | 141.3 | 140.5 | ||||
| 1′ | 25.0 | 0.91 (3H, d, J = 6.0 Hz) | 3′,4′ | 22.7 | 0.91 (3H, d, J = 6.0 Hz) | 2′,3′,4′ |
| 2′ | 21.7 | 0.87 (3H, d, J = 6.0 Hz) | 3′,4′ | 22.6 | 0.89 (3H, d, J = 6.0 Hz,) | 1′,3′,4′ |
| 3′ | 23.1 | 1.89 (1H, t, J = 6.0 Hz) | 4′ | 23.8 | 1.43(1H, m) | 1′,2′,4′ |
| 4′ | 40.1 | 1.66 (1H, t, J = 12.6 Hz) 1.36 (1H, unresolved multiplet) | 1′,2′,3′ | 39.5 | 1.66 (1H, m, overlap) 1.36 (1H, m, overlap) | 3,1′,2′,3′,5′ |
| 5′ | 48.0 | 4.19 (1H, unresolved multiplet) | 4′,7′ | 48.2 | 4.17 (1H, m) | 3′,4′,7′ |
| 6′-NH | 7.99 (1H, d, J = 3.0 Hz) | 4′,5′,7′,8′ | 7.67 (1H, d, J = 9.6 Hz) | 5′,7′ | ||
| 7′ | 171.7 | 170.6 | ||||
| 8′ | 68.9 | 4.01 (1H, unresolved multiplet) | 7′,9′,10′ | 72.01 | 4.12 (1H, d, J = 3.0 Hz) | 7′,9′ |
| 8′-OH | 5.52 (1H, unresolved multiplet)) | 7′,8′,9′ | Unidentified | |||
| 9′ | 76.4 | 3.91 (1H,unresolved multiplet) | 7′,8′,11′ | 85.3 | 4.39 (1H, t, J = 3.0 Hz) | 7′,8′,10′,12′ |
| 10′ | 64.9 | 3.48 (1H, unresolved multiplet) | 12′,2″,5″ | 51.6 | 4.54(1H, dt, J = 10.2 Hz) | 5″,8′,9′,11′,12′ |
| 11′ | 36.4 | 2.23 (1H, d, J = 11.4 Hz) 2.19 (1H, t, J = 12.0 Hz) | 9′,10′,12′ | 34.9 | 2.61 (1H, dd, J = 10.2, 18.0 Hz) 2.18 (1H, dd, J = 10.2, 18.0 Hz) | 9′,10′,12′ |
| 12′ | 172.6 | 175.5 | ||||
| 12′-NH2 | 7.33 (1H, s) 6.87 (1H, s) | 11′,12′ | Unidentified | |||
| 1″ | 167.8 | 166.2 | ||||
| 2″ | 61.8 | 3.28 (1H, unresolved multiplet) | 10′,3″,7″ | 56.8 | 4.04 (1H, t, J = 7.0 Hz) | 10′,1″,5″,7″,8″ |
| 5″ | 107.1 | 170.7 | ||||
| 5″-OH | 9.66 (1H, s) | Unidentified | ||||
| 6″ | 29.0 | 1.28 (3H, s) | 23.3 | 2.07 (3H, s) | 2″,5″ | |
| 7″ | 42.4 | 1.36 (1H, t, J = 6.0 Hz) 1.27 (1H, s, overlap) | 2″,3″,8″ | 39.6 | 1.45 (1H, m) 1.35 (1H, m) | 2″,8″,9″ |
| 8″ | 24.2 | 1.89 (1H, t, J = 6.0 Hz) | 2″,7″,9″,10″ | 24.2 | 1.44 (1H, m, overlap) | 2″,7″,9″,10″ |
| 9″ | 22.7 | 0.82 (3H, d, J = 6.0 Hz) | 7″,8″,10″ | 22.3 | 0.890 (3H, d, J = 6.6 Hz) | 8″,10″ |
| 10″ | 23.4 | 0.79 (3H, d, J = 6.0 Hz) | 7″,8″,9″ | 21.6 | 0.825 (3H, d, J = 6.6 Hz) | 8″,9″ |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, H.-L.; Sun, C.-H.; Hu, X.-X.; You, X.-F.; Wang, M.; Liu, S.-W. Damxungmacin A and B, Two New Amicoumacins with Rare Heterocyclic Cores Isolated from Bacillus subtilis XZ-7. Molecules 2016, 21, 1601. https://doi.org/10.3390/molecules21111601
Tang H-L, Sun C-H, Hu X-X, You X-F, Wang M, Liu S-W. Damxungmacin A and B, Two New Amicoumacins with Rare Heterocyclic Cores Isolated from Bacillus subtilis XZ-7. Molecules. 2016; 21(11):1601. https://doi.org/10.3390/molecules21111601
Chicago/Turabian StyleTang, Hui-Ling, Cheng-Hang Sun, Xin-Xin Hu, Xue-Fu You, Min Wang, and Shao-Wei Liu. 2016. "Damxungmacin A and B, Two New Amicoumacins with Rare Heterocyclic Cores Isolated from Bacillus subtilis XZ-7" Molecules 21, no. 11: 1601. https://doi.org/10.3390/molecules21111601







