Development of a Large Set of Microsatellite Markers in Zapote Mamey (Pouteria sapota (Jacq.) H.E. Moore & Stearn) and Their Potential Use in the Study of the Species
Abstract
:1. Introduction
2. Results and Discussion
2.1. Development of Microsatellite Markers
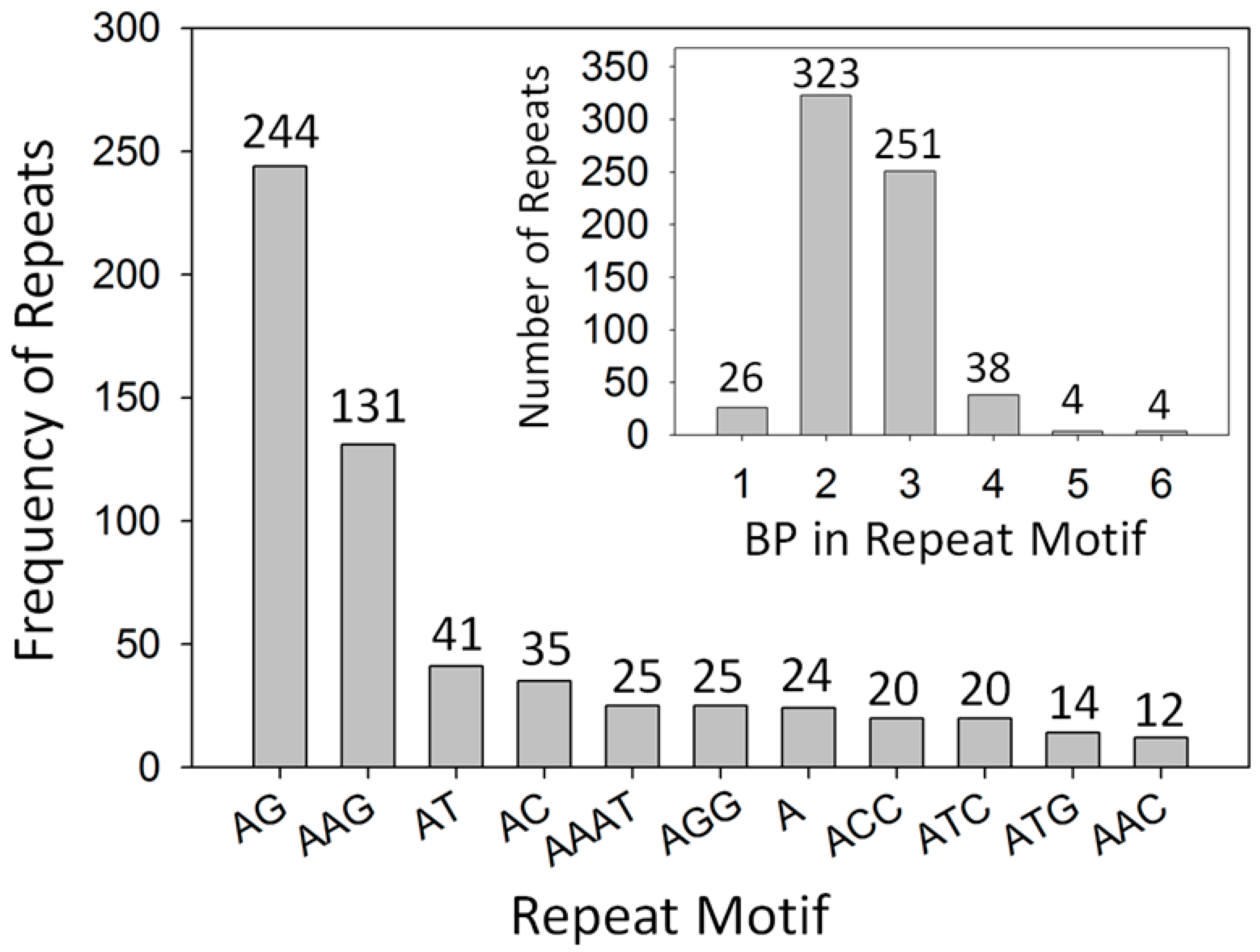
2.2. Genetic Diversity and Founder Effect
| Gene Pools | N | A | HO ± SD | HE ± SD | G–W ± SD |
|---|---|---|---|---|---|
| Wild | 20 | 2.98 | 0.32 ± 0.25 | 0.35 ± 0.20 | 0.49 ± 0.31 |
| Cultivated | 9 | 3.00 | 0.39 ± 0.24 | 0.41 ± 0.17 | 0.42 ± 0.32 |
| All Mexico | 29 | 2.99 | 0.31 ± 0.24 | 0.34 ± 0.21 | 0.46 ± 0.31 |
2.3. Genetic Structure and Cluster Analysis
| Populations | AA | JC | NP | PJ | UR | WH | YJ | AT | DZ | OX |
|---|---|---|---|---|---|---|---|---|---|---|
| AA | *** | 0.11 | 0.03 | 0.03 | 0.11 | 0.03 | 0.06 | 0.04 | 0.05 | 0.11 |
| JC | 3.89 | *** | 0.11 | 0.16 | 0.18 | 0.15 | 0.12 | 0.13 | 0.14 | 0.22 |
| NP | 14.63 | 3.75 | *** | 0.01 | 0.10 | 0.04 | 0.02 | 0.06 | 0.08 | 0.14 |
| PJ | 13.70 | 2.61 | 74.10 | *** | 0.07 | 0.02 | 0.07 | 0.03 | 0.05 | 0.12 |
| UR | 3.93 | 2.25 | 4.40 | 5.90 | *** | 0.12 | 0.02 | 0.10 | 0.14 | 0.21 |
| WH | 12.20 | 2.79 | 9.71 | 17.98 | 3.61 | *** | 0.07 | 0.07 | 0.12 | 0.16 |
| YJ | 6.72 | 3.42 | 21.95 | 5.97 | 23.38 | 6.57 | *** | 0.08 | 0.13 | 0.18 |
| AT | 12.00 | 3.21 | 7.79 | 15.18 | 4.10 | 6.16 | 5.44 | *** | 0.08 | 0.06 |
| DZ | 8.42 | 2.92 | 5.68 | 8.11 | 2.98 | 3.52 | 3.16 | 5.64 | *** | 0.06 |
| OX | 3.95 | 1.75 | 2.99 | 3.55 | 1.87 | 2.53 | 2.14 | 7.06 | 6.92 | *** |
| Source of Variation | Degree of Freedom | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among groups (wild and domesticated gene pools) | 1 | 59.848 | 0.99087 Va | 3.82 |
| Among populations within groups | 8 | 279.256 | 2.08706 Vb | 8.05 |
| Within populations | 48 | 1097.000 | 22.85417 Vc | 88.13 |
| Total | 57 | 1436.103 | 25.93210 |
 : distance between two locations in kilometers, not at scale.
: distance between two locations in kilometers, not at scale.
 : distance between two locations in kilometers, not at scale.
: distance between two locations in kilometers, not at scale.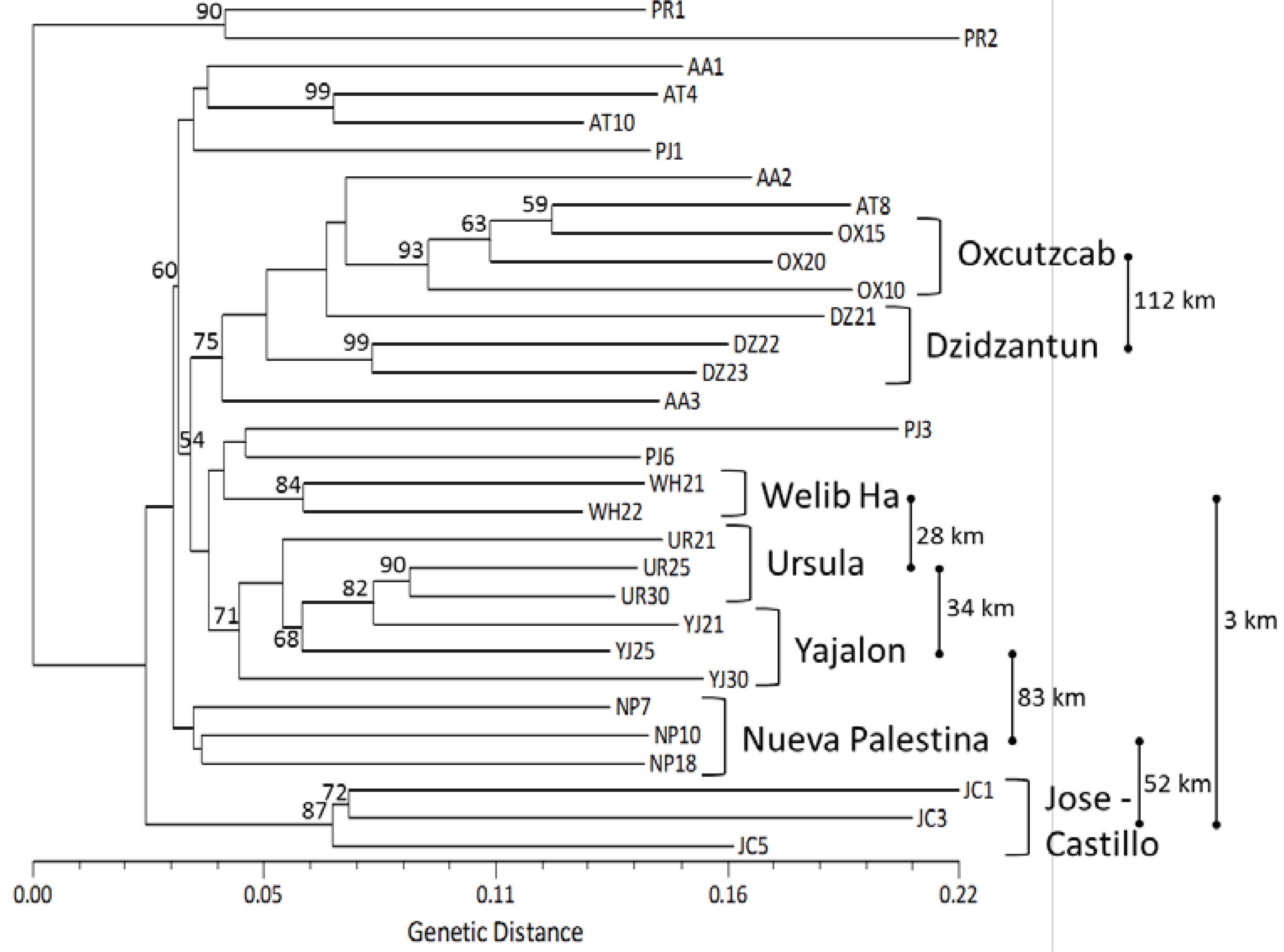
| ID | Origin | Accession | Location | Coordinates |
|---|---|---|---|---|
| PR1 | cultivated | Lorito #2; TARS: 17854 | B-IV, T-2 F-1, Isabela, Puerto Rico | 18°30′47″ N 67°04′12″ W |
| PR2 | Cultivated | Pace; TARS: 17866 | B-IV, T-2, F-13, Isabela, Puerto Rico | |
| AA1 | wild | (J)_1 | Agua Azul, Mexico | 17°15.494′ N 92°06.824′ W |
| AA2 | wild | (J)_2 | Agua Azul, Mexico | |
| AA3 | wild | (J)_3 | Agua Azul, Mexico | |
| JC1 | wild | (E)_1 | Jose Castillo, Mexico | 17°23.807′ N 91°48.748′ W |
| JC3 | wild | (E)_3 | Jose Castillo, Mexico | |
| JC5 | wild | (E)_5 | Jose Castillo, Mexico | |
| NP7 | wild | (L)_7 | Nueva Palestina, Mexico | 16°57.810′ N 91°37.665′ W |
| NP10 | wild | (L)_10 | Nueva Palestina, Mexico | |
| NP18 | wild | (L)_18 | Nueva Palestina, Mexico | |
| PJ1 | wild | (K)_1 | Penjamo, Mexico | 17°26.041′ N 91°37.665′ W |
| PJ3 | wild | (K)_3 | Penjamo, Mexico | |
| PJ6 | wild | (K)_6 | Penjamo, Mexico | |
| UR21 | wild | (B)_21 | Ursula, Mexico | 17°18.416′ N 92°02.997′ W |
| UR25 | wild | (B)_25 | Ursula, Mexico | |
| UR30 | wild | (B)_30 | Ursula, Mexico | |
| WH21 | wild | (C)_21 | Welib Ha, Mexico | 17°22.521′ N 91°47.893′ W |
| WH22 | wild | (C)_22 | Welib Ha, Mexico | |
| YJ21 | wild | (A)_21 | Yajalon, Mexico | 17°13.475′ N 92°21.184′ W |
| YJ25 | wild | (A)_25 | Yajalon, Mexico | |
| YJ30 | wild | (A)_30 | Yajalon, Mexico | |
| AT4 | Cultivated | (G)_4 | Atasta, Mexico | 18°36′58.9″ N 92°9′12.13″ W |
| AT8 | Cultivated | (G)_8 | Atasta, Mexico | |
| AT10 | Cultivated | (G)_10 | Atasta, Mexico | |
| DZ21 | Cultivated | (M)_21 | Dzidzantun, Mexico | 21°14′53.5″ N 89°1′59.65″ W |
| DZ22 | Cultivated | (M)_22 | Dzidzantun, Mexico | |
| DZ23 | Cultivated | (M)_23 | Dzidzantun, Mexico | |
| OX10 | Cultivated | (F)_10 | Oxcutzcab, Mexico | 20°18′10″ N 89°25′0″ W |
| OX15 | Cultivated | (F)_15 | Oxcutzcab, Mexico | |
| OX20 | Cultivated | (F)_20 | Oxcutzcab, Mexico |
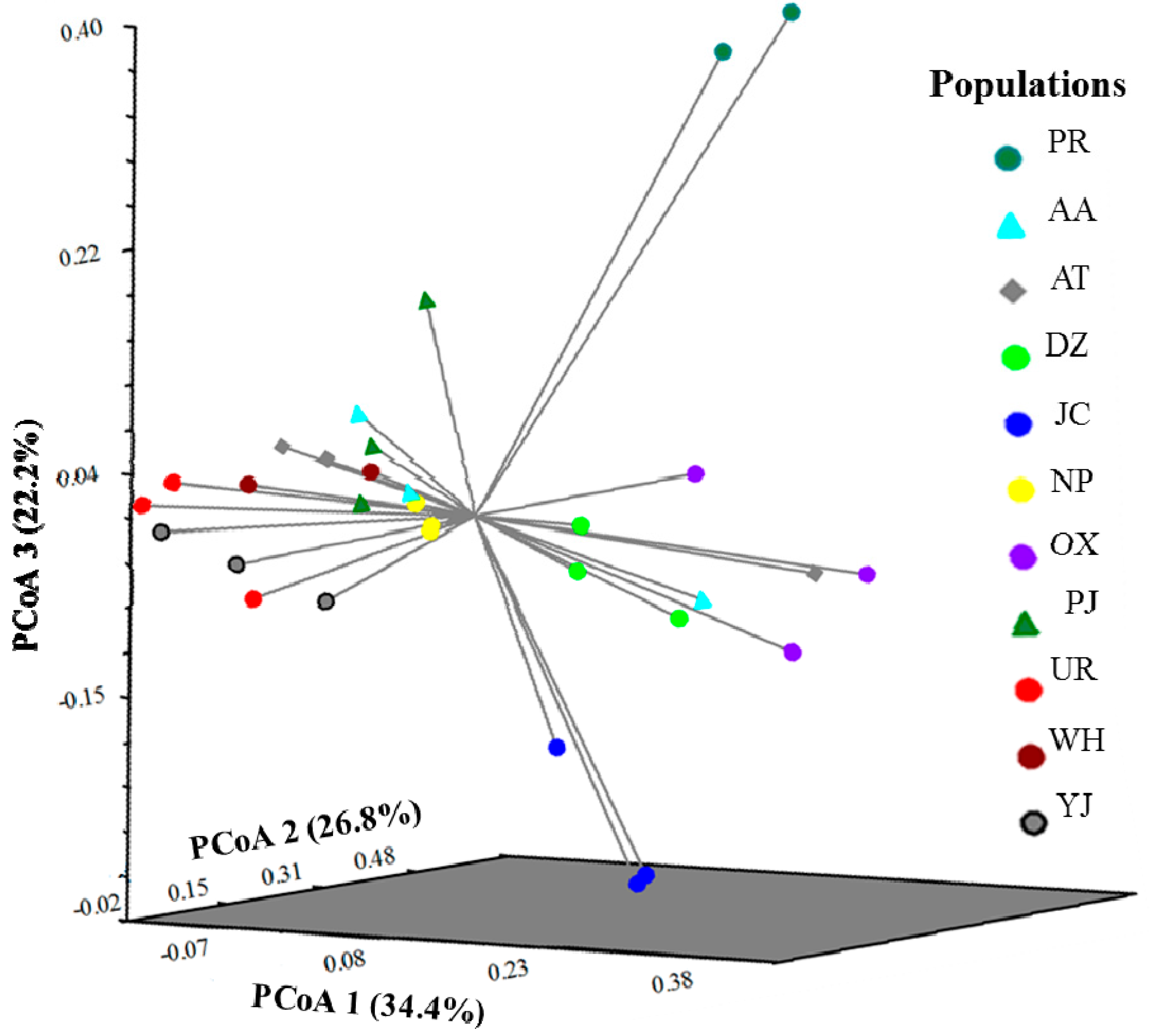
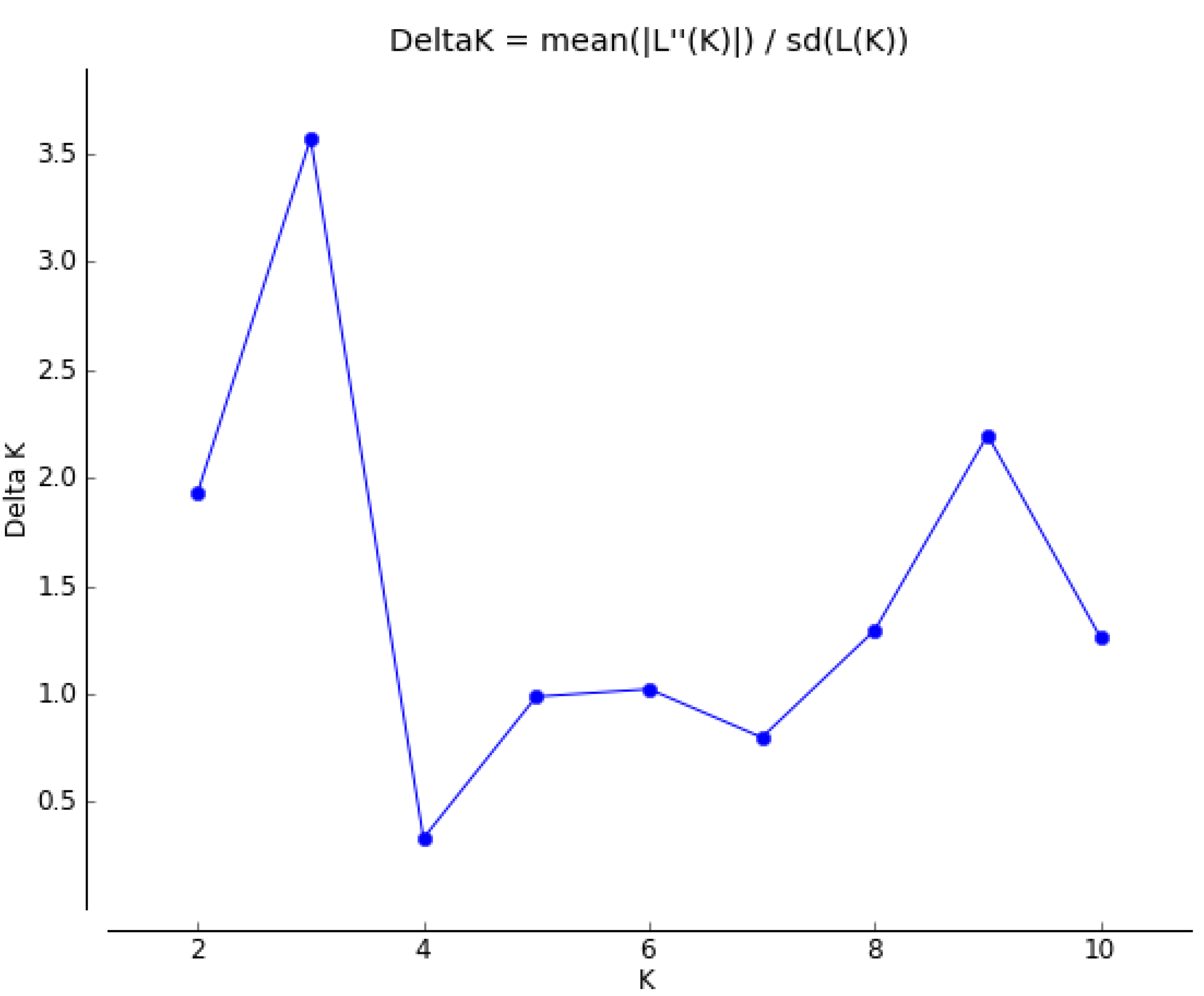

3. Experimental Section
3.1. Plant Material, DNA Extraction, and Isolation of Microsatellites
3.2. Fingerprinting
3.3. Genetic Diversity and Cluster Analyses
3.4. Genetic Structure and Cluster Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bartish, I.V.; Swenson, U.; Munzinger, J.R.M.; Anderberg, A.A. Phylogenetic relationships among New Caledonian Sapotaceae (Ericales): Molecular evidence for generic polyphyly and repeated dispersal. Am. J. Bot. 2005, 92, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Triono, T.; Brown, A.H.D.; West, J.G.; Crisp, M.D. A phylogeny of Pouteria (Sapotaceae) from Malesia and Australasia. Aust. Syst. Bot. 2007, 20, 107–118. [Google Scholar] [CrossRef]
- Morera, J.A. El Zapote; CATIE/GTZ: Turrialba, Costa Rica, 1982; p. 24. [Google Scholar]
- Pennington, T.D. Flora Neotropica. Monograph 52. Sapotaceae; New York Botanical Garden Press: New York, NY, USA, 1990; p. 770. [Google Scholar]
- Azurdia, C. Tres Especies de Zapote en America Tropical; Southampton University: Southampton, UK, 2006; p. 216. [Google Scholar]
- Bruner, B.R.; Morales-Payan, J.P. Soils, Plant Growth and Crop Production—Sapote, Sapodilla and Star Apple. Available online: http://www.eolss.net/sample-chapters/c10/e1-05a-53-00.pdf (accessed on 10 April 2015).
- SIAP-SAGARPA. Anuario Estadístico de la Producción de los Estados Unidos Mexicanos; Secretaría de Agricultura; Ganadería y Desarrollo Rural: México, 2010; Available online: www.siap.gob.mx (accessed on 20 April 2015).
- Gulyas-Fekete, G.; Murillo, E.; Kurtan, T.; Papp, T.; Illyés, T.-Z.; Drahos, L.; Visy, J.; Agócs, A.; Turcsi, E.; Deli, J. Cryptocapsinepoxide-Type Carotenoids from Red Mamey, Pouteria sapota. J. Nat. Prod. 2013, 76, 607–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murillo, E.; McLean, R.; Britton, G.; Agócs, A.; Nagy, V.; Deli, J. Sapotexanthin, an A-Provitamin Carotenoid from Red Mamey (Pouteria sapota). J. Nat. Prod. 2011, 74, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.; Jungfer, E.; da Silva, B.A.; Maia, J.G.S.; Marx, F. Phenolic constituents and antioxidant capacity of four underutilized fruits from the Amazon region. J. Agric. Food Chem. 2011, 59, 7688–7699. [Google Scholar] [CrossRef] [PubMed]
- Moo-Huchin, V.; Estrada-Mota, I.; Estrada-León, R.; Cuevas-Glory, L.F.; Sauri-Duch, E. Chemical composition of crude oil from the seeds of pumpkin (Cucurbita spp.) and mamey sapota (Pouteria sapota Jacq.) grown in Yucatan, Mexico. CyTA—J. Food. 2013, 11, 324–327. [Google Scholar] [CrossRef]
- Morton, J.F. Sapotaceae. In Fruits of Warm Climates; Morton, J.F., Ed.; Julia F. Morton: Miami, FL, USA, 1987; pp. 398–402. [Google Scholar]
- Silva, C.A.M.; Simeoni, L.A.; Silveira, D. Genus Pouteria: Chemistry and biological activity. Rev. Bras. Farmacogn. 2009, 19, 501–509. [Google Scholar] [CrossRef]
- Miller, A.J.; Schaal, B.A. Domestication and the distribution of genetic variation in wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea L. (Anacardiaceae). Mol. Ecol. 2006, 15, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Gross, B.L. From forest to field: Perennial fruit crop domestication. Am. J. Bot. 2011, 98, 1389–1414. [Google Scholar] [CrossRef] [PubMed]
- Espinos-Zaragoza, S.; Saucedo, V.C.; Villegas, M.A.; Ibarra, E.M.E. Caracterización de frutos de zapote mamey (Pouteria sapota (Jacq.) H.E. Moore & Stearn) en Guerrero, México. Proc. Interamer. Soc. Trop. Hort. 2005, 48, 135–138. [Google Scholar]
- Bañuelos-Jimenez, J.S.; Ochoa, I. Caracterización morfológica de zapote mamey (Pouteria sapota Jacq. H.E. Moore & Stearn) del centro occidente de Michoacán, México. Rev. Fitotec. Mex. 2006, 29, 9–17. [Google Scholar]
- Gaona-García, A.; Alia-Tejacal, I.; López-Martínez, V.; Andrade-Rodríguez, M.; Colinas-León, M.T.; Villegas-Torres, O. Caracterización de frutos de zapote mamey (Pouteria sapota) en el suroeste del estado de Morelos. Rev. Chapingo Serie Hortic. 2008, 14, 41–47. [Google Scholar]
- Nascimento, V.E.; Martins, A.B.G.; Hojo, R.H. Caracteizaçao física e química de fruto de mamey. Rev. Bras. Frut. 2008, 30, 953–957. [Google Scholar] [CrossRef]
- Carrara, S.; Campbell, R.; Schnell, R. Genetic variation among cultivated selections of mamey sapote (Pouteria spp. [Sapotaceae]). Proc. Fla. State Hort. Soc. 2004, 117, 195–200. [Google Scholar]
- Ibarra-Estrada, M.E. Morfología de Frutos, Perfil de Ácidos Grasos en Semilla y Diversidad Genética Molecular en Selecciones de Zapote Mamey [Pouteria sapota (Jacq.) H.E. Moore & Stearn]. Master Thesis, Colegio de Postgraduados, Campus Montecillo, México, April 2012. [Google Scholar]
- Rodríguez-Rojas, T.J.; Andrade-Rodríguez, M.; Alia-Tejacal, I.; López-Martínez, V.; Espinosa-Zaragoza, S.; Esquinca-Avilés, H. Caracterización molecular de zapote mamey [Pouteria sapota (Jacq.) Moore & Stearn]. Rev. Fac. Agron. (LUZ) 2012, 29, 339–354. [Google Scholar]
- Glenn, T.C.; Schable, N.A. Isolating microsatellite DNA loci. Method Enzymol. 2005, 395, 202–222. [Google Scholar]
- Techen, N.; Arias, R.S.; Pan, Z.; Khan, I.A.; Scheffler, B.E. Optimized construction of microsatellite-enriched libraries. Mol. Ecol. Res. 2010, 10, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Kalia, R.K.; Singh, R.; Rai, M.K.; Dhawan, A.K.; Kalia, S. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Arias, R.S.; Ballard, L.L.; Scheffler, B.E. UPIC: Perl scripts to determine the number of SSR markers to run (Software). Bioinformation 2009, 3, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Kanashiro, M.; Ciampi, A.; Grattapagua, D. Genetic structure and mating system of Manilkara huberi (Ducke) A. Chew., a heavily logged Amazonian timber species. J. Hered. 2007, 98, 646–654. [Google Scholar] [CrossRef] [PubMed]
- González-Hernández, D.; García-Pérez, E.; Guntin-Marey, P. Genetic characterization of Manilkara zapota from Veracruz, México, with SSR markers. Agrociencia 2012, 46, 663–675. [Google Scholar]
- Majourhat, K.; Jabbar, Y.; Hadi, A.; Martínez-Gomez, P. Molecular characterization and genetic relationships among most common identied morphotypes of critically endangered rare Moroccan species Argania spinosa (Sapotaceae) using RAPD and SSR markers. Ann. For. Sci. 2008, 65. [Google Scholar] [CrossRef]
- Toledo, C.L. Efecto de la Fragmentación en la Estructura Genética y en Características de Historias de vida de Sideroxylon Portoricense (Sapotaceae) en la Selva de los Tuxtlas Veracruz, México. Bachelor Thesis, Facultad de Ciencias, Universidad Nacional Autónoma de México, México, May 2009. [Google Scholar]
- Pickersgill, B. Cytological and genetical evidence on the domestication and diffusion of crops within the Americas. In Foraging and Farming: The Evolution of Plant Exploitation; Harris, D.R., Hillman, G.C., Eds.; Unwin Hyman: London, UK, 1989; pp. 426–439. [Google Scholar]
- Zohary, D. Domestication of the carob (Ceratonia siliqua L.). Isr. J. Plant Sci. 2002, 50, S141–S145. [Google Scholar] [CrossRef]
- De Baan, L.; Mutel, C.L.; Curran, M.; Hellweg, S.; Koellner, T. Land use in life cycle assessment: Global characterization factors based on regional and global potential species extinction. Environ. Sci. Technol. 2013, 47, 9281–9290. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.M.; Fowler, J.C.; Garbett, C.A.; Thomas, M.R. Enrichment of microsatellites from the citrus genome using biotinylated oligonucleotide sequences bound to streptavidin-coated magnetic particles. Biotechniques 1994, 16, 656–660. [Google Scholar] [PubMed]
- Sharopova, N.; McMullen, M.D.; Schultz, L.; Schroeder, S.; Sanchez-Villeda, H.; Gardiner, J.; Bergstrom, D.; Houchins, K.; Melia-Hancock, S.; Musket, T.; et al. Development and mapping of SSR markers for maize. Plant Mol. Biol. 2002, 48, 463–481. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Bioinformatics Methods and Protocols in the Series Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2000. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Waldbieser, G.C.; Quiniou, S.M.; Karsi, A. Rapid development of gene-tagged microsatellite markers from bacterial artificial chromosome clones using anchored TAA repeat primers. Biotechniques 2003, 35, 976–979. [Google Scholar] [PubMed]
- Brownstein, M.J.; Carpten, J.D.; Smith, J.R. Modulation on non-templated nucleotide addition by Taq DNA polymerase: Primers modifications that falicilitate genotyping. Biotechniques 1996, 20, 1004–1006, 1008–1010. [Google Scholar] [PubMed]
- Kalinowski, S.T. HP-Rare: A computer program for performing rarefaction on measures of allelic diversity. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin version 3.0: An integrated software package for population genetics data analysis. Evolut. Bioinform. Online 2005, 1, 47–50. [Google Scholar]
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Luikart, G.; Cornuet, J.M. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 1997, 12, 228–237. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.; Quattro, J. Analysis of molecular variance inferred from metric distance among DNA haplotypes: Applications to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [PubMed]
- Slatkin, M.A. Measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Sokal, R.R. Testing statistical significance of geographic variation patterns. Syst. Zool. 1979, 28, 227–232. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Efron, B.; Halloran, E.; Holmes, S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 1996, 93, 13429–13434. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–391. [Google Scholar] [CrossRef]
- Yap, I.V.; Nelson, R.J. Winboot: A Program for Performing Bootstrap Analysis of Binary Data to Determine the Confidence limits of UPGMA Based Dendrograms; IRRI Discussion Paper Series No. 14; International Rice Research Institute: Manila, Philippines, 1996; Available online: https://sites.google.com/site/pbg804/winboot (accessed on 18 June 2015).
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.1; Exeter Publishing: Setauket, NY, USA, 2001. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- National Plant Germplasm System, Germplasm Resources Information Network (GRIN). Available online: http://www.ars-grin.gov/npgs/holdings.html (accessed on 18 June 2015).
- JGI—GOLD, Genomes On Line Database. Available online: https://gold.jgi-psf.org (accessed on 18 June 2015).
- Jessup, L.W.; Short, P.S. Flora of the Darwin Region, Volume 1—Sapotaceae; Northern Territory Government: Palmerston, Australia, 2011.
- Heer, K.; Machado, C.A.; Himler, A.G.; Herre, E.A.; Kalko, E.K.V.; Dick, C.W. Anonymous and EST-based microsatellite DNA markers that transfer broadly across the fig tree genus (Ficus, Moraceae). Am. J. Bot. 2012, 99, e330–e333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrede, I.; Carlsen, T.; Rieseberg, L.; Brochmann, C. Microsatellites for three distantly related genera in the Brassicaceae. Conserv. Genet. 2009, 10, 643–648. [Google Scholar] [CrossRef]
- Arias, R.S.; Ray, J.D.; Mengistu, A.; Scheffler, B.E. Discriminating microsatellites from Macrophomina phaseolina and their potential association to biological functions. Plant Pathol. 2011, 60, 709–718. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the plant material used in this study are available from the first author.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arias, R.S.; Martínez-Castillo, J.; Sobolev, V.S.; Blancarte-Jasso, N.H.; Simpson, S.A.; Ballard, L.L.; Duke, M.V.; Liu, X.F.; Irish, B.M.; Scheffler, B.E. Development of a Large Set of Microsatellite Markers in Zapote Mamey (Pouteria sapota (Jacq.) H.E. Moore & Stearn) and Their Potential Use in the Study of the Species. Molecules 2015, 20, 11400-11417. https://doi.org/10.3390/molecules200611400
Arias RS, Martínez-Castillo J, Sobolev VS, Blancarte-Jasso NH, Simpson SA, Ballard LL, Duke MV, Liu XF, Irish BM, Scheffler BE. Development of a Large Set of Microsatellite Markers in Zapote Mamey (Pouteria sapota (Jacq.) H.E. Moore & Stearn) and Their Potential Use in the Study of the Species. Molecules. 2015; 20(6):11400-11417. https://doi.org/10.3390/molecules200611400
Chicago/Turabian StyleArias, Renée S., Jaime Martínez-Castillo, Victor S. Sobolev, Nasib H. Blancarte-Jasso, Sheron A. Simpson, Linda L. Ballard, Mary V. Duke, Xiaofen F. Liu, Brian M. Irish, and Brian E. Scheffler. 2015. "Development of a Large Set of Microsatellite Markers in Zapote Mamey (Pouteria sapota (Jacq.) H.E. Moore & Stearn) and Their Potential Use in the Study of the Species" Molecules 20, no. 6: 11400-11417. https://doi.org/10.3390/molecules200611400






