S4MPLE—Sampler for Multiple Protein-Ligand Entities: Methodology and Rigid-Site Docking Benchmarking
Abstract
:1. Introduction
- rigid docking (target and ligand are considered rigid)
- semi-flexible docking (flexible ligand and rigid target)
- flexible docking (flexible ligand and partly flexible target)
1.1. Sampling Strategies
1.2. Scoring Functions
1.3. Overview of This Article
- The key novelty in S4MPLE: genetic operators supporting the unified handling of both intra- and inter-molecular DoF
- Differentiable interaction fingerprints to monitor conformer diversity—another important original development
- Employed evolutionary strategies, the originality of which draws on the novelty of involved operators and diversity control tools
- The set-up and fitting of the force field (FF) engine used for energy calculations, based on the classical AMBER [39,40]/Generalized AMBER (GAFF) [39,41]. The introduction of classical FF terms is combined with the introduction of—occasionally innovative, often adapted—additional terms: chirality control terms, a continuum solvent model and favorable contact (hydrophobic, H bonding) bonus terms. The latter, together with classical customizable FF terms—such as the chosen dielectric constant, and a herein introduced repulsive van der Waals coefficient weighing term—need fitting.
- Introduction of the herein used data sets.
- The fitting protocol of the additional terms. This may count as a conceptual novelty in as far as calibration efforts based on published ligand-protein complexes of known affinities were so far reserved to derivations of free energy-predicting scoring functions [7,42], not to fine-tune the FF engine per se.
- Eventually, the redocking protocol used to asses S4MPLE proficiency in classical docking benchmarks is given, in parallel to the similar protocol running FlexX, for comparison purposes.
2. Experimental Section
2.1. S4MPLE
2.2. Hybrid Evolutionary Operators
2.2.1. Atom Flexibility Status
2.2.2. Cross-Over Operators
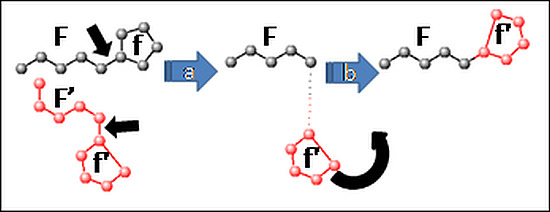
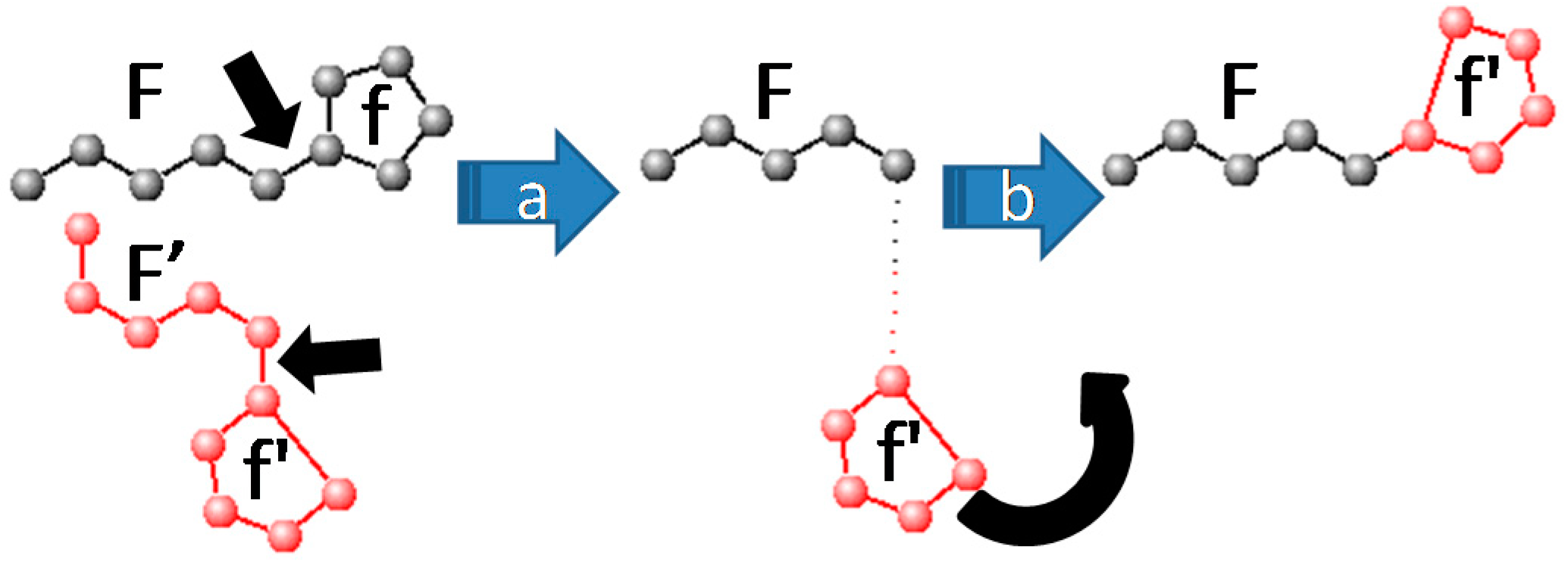
- Fragment recombination randomly picks a pair of complementary molecular fragments (F, f) defined at the beginning of the simulation. Such fragments may correspond either to two radicals (F containing more atoms than f), covalently connected to each other by a rotatable bond (see Figure 1), or a loose ligand f moving freely with respect to the receptor moiety F. In this way, folding and docking, the two key applications typically dealt with by different software suites, are here unified. The fragment recombination takes place in several stages. First (step a in Figure 1) the geometry of F (xij, atom i∈F, coordinate j = 1–3) is taken from the first mate, whereas the geometry of f is imported from the second, as f'. Next (step b), f' will be reconnected to F, i.e. the imported coordinates (xij, i∈f') are submitted to a roto-translation meant to replace them in a chemically meaningful way with respect to F.
- ○
- DockC randomly rotates f' around the F–f' axis, either until a clash-free arrangement is obtained, or a maximal number of attempts (50) is reached.
- ○
- DockE pursues random rotations while attempting to minimize the inter-fragment interaction energies. It stops and returns the most stable pose found so far if 20 successive attempts failed to discover any better one.
- Uniform torsional crossovers browse through the list of rotatable bonds, randomly pick one of the two parents as “donor” and set the associated torsion angle value in the child geometry equal to the one of the donor. This is a usual type of crossing-over operator. LO is then used to tentatively relax the atom clashes in the brute geometry associated to the combined list of torsion angle values.
2.2.3. Mutations
2.2.4. ‘Lamarckian’ Local Optimization
2.2.5. Exhaustive Energy Minimization
2.2.6. Initialization
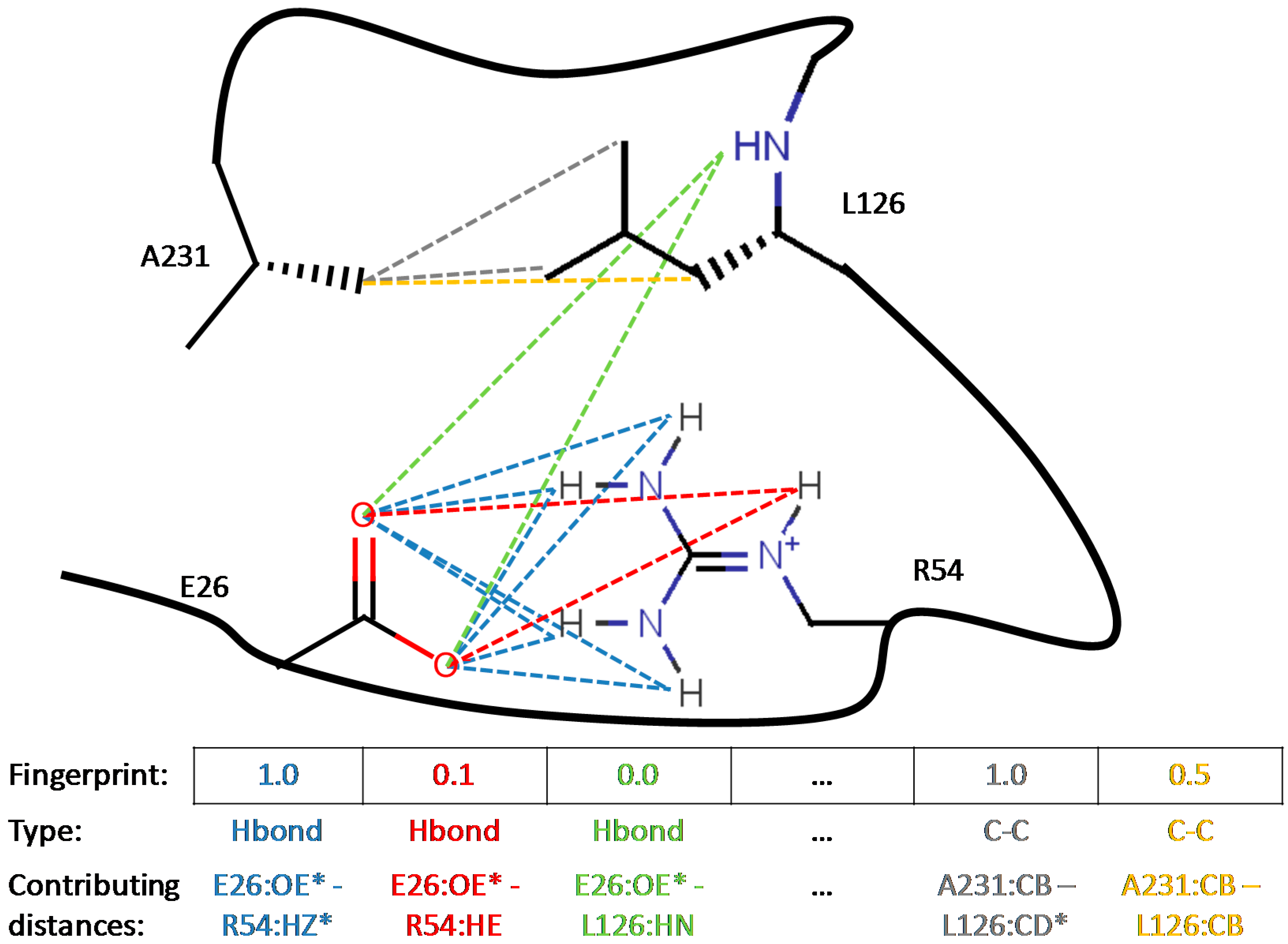
2.3. Population Diversity Control: Interaction Fingerprints
- general, regrouping both intra- and intermolecular favorable contacts: hydrogen bonds and hydrophobic contacts
- symmetry-compliant, i.e. invariant to swapping of the contact status of topologically equivalent atoms: rotation of 180° of a carboxylate group having one of its oxygens acting as acceptor in a hydrogen bond will not change the fingerprint, as the hydrogen bond now involves a different, yet topologically equivalent oxygen.
- differentiable, rather than binary: contact status varies smoothly between 0 (absent) and 1 (fully established) as the corresponding contact distance scans [dmin, dmax].
- H-bond donors : H, HO, HW, hn, ho, hw
- H-bond acceptors : O, O2, OW, OH, OS, NB, NC, NY, o, oh, os, ow, n1, n2, n3, na, nb, nc, nd, ne, nf
| Category | Force Field | Atomic Type | Description (from AMBER/GAFF Parameters File) |
|---|---|---|---|
| Polarized | AMBER | C | sp2 C carbonyl group |
| GAFF | c | sp2 C carbonyl group | |
| Aromatic | AMBER | C* | sp2 arom. 5 memb.ring w/1 subst. (TRP) |
| AMBER | CA | sp2 C pure aromatic (benzene) | |
| AMBER | CB | sp2 aromatic C, 5&6 membered ring junction | |
| AMBER | CC | sp2 aromatic C, 5 memb. ring HIS | |
| AMBER | CN | sp2 C aromatic 5&6 memb.ring junct.(TRP) | |
| AMBER | CR | sp2 arom as CQ but in HIS | |
| AMBER | CV | sp2 arom. 5 memb.ring w/1 N and 1 H (HIS) | |
| AMBER | CW | sp2 arom. 5 memb.ring w/1 N-H and 1 H (HIS) | |
| GAFF | ca | sp2 C in pure aromatic systems | |
| GAFF | cc | sp2 carbons in non-pure aromatic systems | |
| GAFF | cd | sp2 carbons in non-pure aromatic systems | |
| GAFF | cp | Head sp2 carbons connecting rings in bi-phenyls | |
| GAFF | cq | Head sp2 carbons connecting rings in bi-phenyls | |
| Aliphatic | AMBER | CT | sp3 aliphatic C |
| GAFF | c1 | sp C | |
| GAFF | c2 | sp2 C | |
| GAFF | c3 | sp3 C | |
| GAFF | ce | Inner sp2 carbons in conjugated systems | |
| GAFF | cf | Inner sp2 carbons in conjugated systems | |
| GAFF | cg | Inner sp carbons in conjugated systems | |
| GAFF | ch | Inner sp carbons in conjugated systems | |
| GAFF | cu | sp2 carbons in triangle systems | |
| GAFF | cv | sp2 carbons in square systems | |
| GAFF | cx | sp3 carbons in triangle systems | |
| GAFF | cy | sp3 carbons in square systems |
- first, ωmin = min[ω(M)]—the minimal degree of originality over all members M of the population—is found. MR is taken to be the least fit individual among the least original members M (those with ω(M) ≤ 1.1ωmin).
- An empirical trade-off between diversification and acceptation of less fit individuals is achieved: individual O replaces MR if its excess energy E(O)−E(MR) in kcal/mol is ‘compensated’ by the gain in diversity, at user-defined div2E parameter. If E(O)−E(MR) ≤ div2E[ω(O)−ω(MR)] and ω(O) > ω(MR), then the individual O replaces MR, and an offspring is accepted in the population and coexist with its parents. By default, children only replace their parents.
- M represents an acceptable trade-off between fitness loss vs. originality gain with respect to I: E(M) − E(I) < div2E[ω(M) − ω(I)], or
- the allotted number of attempts (5) is reached.
2.4. Evolutionary Strategies
2.5. Force Field
2.5.1. Continuum Solvent Model and Contact Terms
- A pair-based desolvation term (Equation (3)) [51], function of partial charges Q and distance d between two atoms i and j, and scaled by a generic constant (more about this in the force field fitting paragraph):
- A linearly distance-dependent relative dielectric constant (εr) is used in the Coulomb term [52], which de facto makes it a function of 1/d2:
- Contact terms (Equation (5)) [51], rewarding favorable interactions such as hydrophobic contacts and hydrogen bonds were added to the FF. In this context, hydrophobic contacts refer to close carbons in space, and hydrogen bond donors are hydrogens on heteroatoms. Constant κij is a function of the nature of the contact, and the types of involved atoms i and j, see parameter fitting below. Cij encodes contact strength: full contact Cij = 1 is assumed at dij < dmin. Contact ceases completely at dij > dmax, and its strength varies smoothly within the switching range [dmin, dmax]. This range is contact type dependent: for hydrophobic contacts, a range of [4.5, 5.5] Ǻ is used. For hydrogen bonds, the range is atom pair specific: [(sum of vdW Radii) − 0.5, (sum of vdW Radii) + 0.1]:
2.5.2. Context-Specific Termination Function
2.5.3. Out-of-Plane and Chiral Constraint Terms
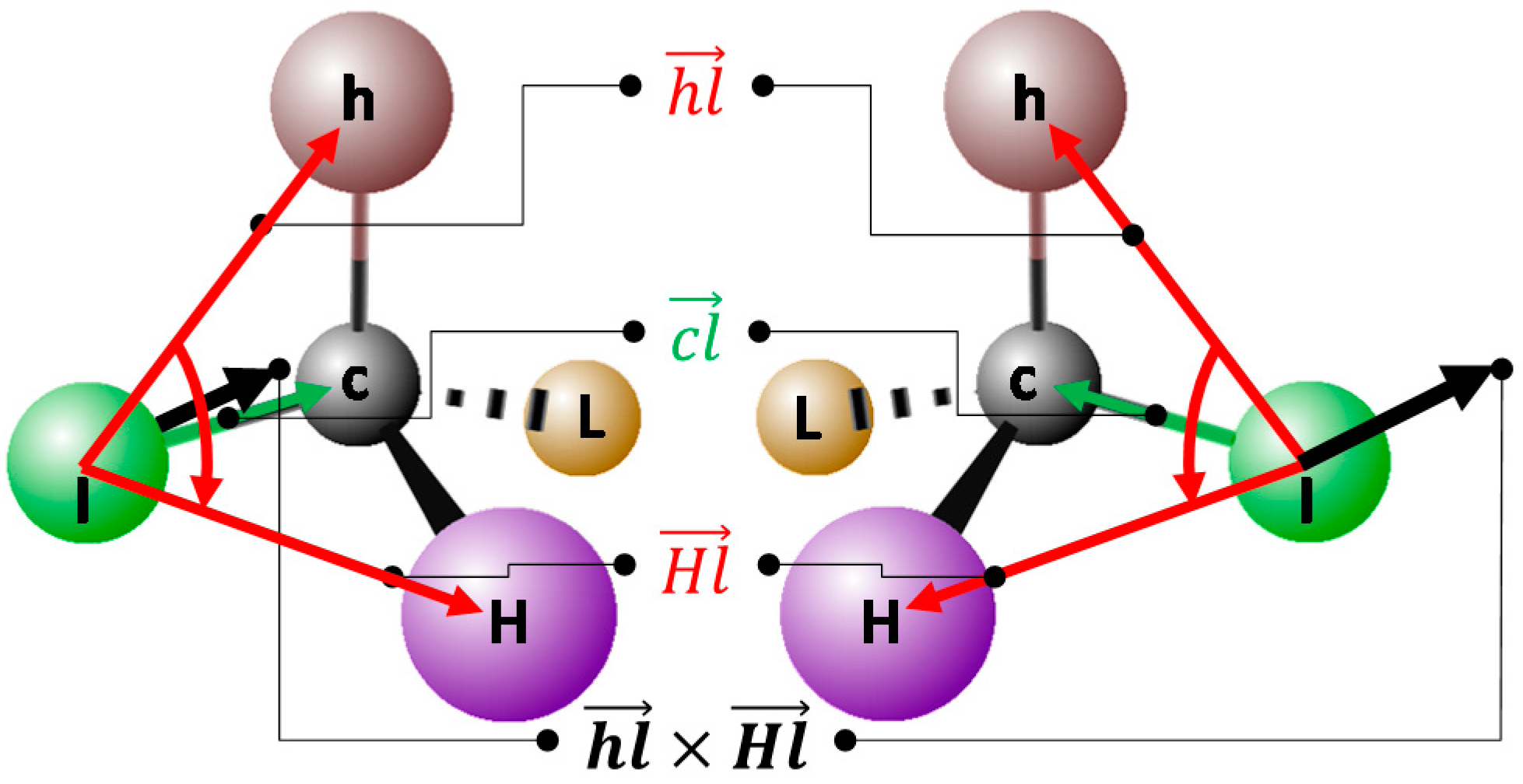
2.5.4. Potential Energy Surface: Avoiding Singularities
2.6. Datasets
2.6.1. Astex/CCDC Clean Subset
- No covalently bound ligand
- No complexes where the symmetry-related units are mandatory to explain the binding mode (as seems to be the case, among other, for 1ETA or 3CLA—check performed using the “symexpˮ command in Pymol [54]).
2.6.2. Astex Diverse Set
2.7. Force Field Parameter Tuning Protocol
- epsilon: proportionality constant of the distance-dependent relative dielectric constant εr = epsilon × d.
- repulsive_factor (default 1.0): the weight of the an der Waals repulsive term. Therefore, , where Aij and Bij represent the AMBER/GAFF vdW pairwise interaction coefficients.
- vicinal_weight: for vicinal (three bonds apart) pairs i, j, the entire vdW term EvdWij is scaled down by vicinal_weight, a trick already employed in the original AMBER.
- desolv_factor : the value to be assigned to the desolvation parameter σ from Equation (3), for all the pairs i, j except the special cases outlined below:
- minq_to_desolv: the minimal charge threshold for the desolvation term (i.e. if max(|Qi|, |Qj|) < minq_to_desolv then = 0).
- desolv_scale_ion and desolv_scale_hb: the value of scaling factors for some specific desolvation terms (pairs involving ion or hydrogen-bonds). These extra parameters were required in order to achieve successful docking predictions—a preliminary attempt to fit a set of parameters including only desolv_factor to control desolvation failed (results not shown).
- hbond_bonus: the common value for all κij in Equation (5) corresponding to hydrogen bond interactions.
- Hydrophobic constants Kc for each hydrophobic carbon class (see Table 1), introduced in order to define hydrophobic contact constants κij in Equation (5) as the average of constants of the involved carbons.
| Parameter | Core FF | Preliminary FF | Fit FF |
|---|---|---|---|
| epsilon | 2 | 4 | 4 |
| desolv_factor | 0.0 | 0.1 | 0.1 |
| minq_to_desolv | 0 | 0.125 | 0.125 |
| hbond_bonus | 0 | 2 | 2 |
| repulsive_factor | 1.00 | 0.75 | 0.75 |
| vicinal_weight | 0.5 | 0.033 | 0.033 |
| desolv_scale_ion | – | 1.0 | 0.1 |
| desolv_scale_hb | – | 1.0 | 0.1 |
| Kpolarized | 0.0 | 0.1 | 0.01 |
| Karom | 0.0 | 0.1 | 0.15 |
| Kaliph | 0.0 | 0.1 | 0.15 |
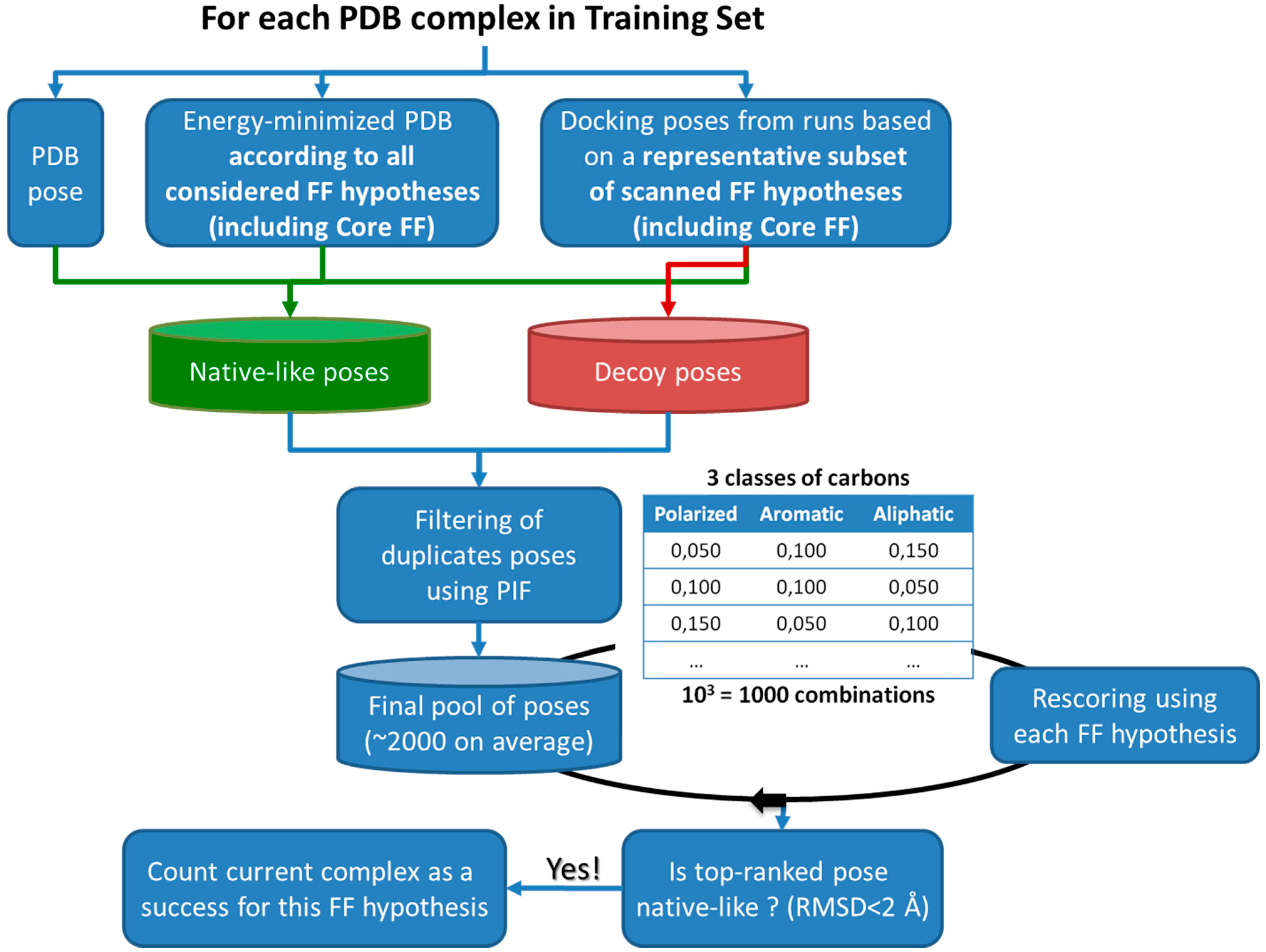
- First, tuning of the desolvation parameters (desolv_scale_ion and desolv_scale_hb), based on the observations that the Preliminary set-up leads to both (a) bad coordination between ligand and metal ion for several metallo-protein complexes (e.g., mono-dentate coordination preferred to bi-dentate coordination for the acid group in 1CBX), and (b) distorted hydrogen bonds for complexes involving sugars. Thus, the idea to specifically rescale desolvation term of the above-cited classes of interactions by means of weights, fixed by trial-and-error docking simulations of concerned protein-ligand complexes.Figure 4. Strategy used in the force field parameter tuning protocol. ‘Decoy poses’ are non-native-like, to be distinguished from the native-like by a proper choice of FF parameters. These three parameters were subjected to a systematic scan. Within the scanned parameter space volume, the representative subset of possible FF configurations includes: Core FF, Preliminary FF setups, setups at the extremes (corners) and at the center of the scanned parameter ‘cube’. This representative subset is used to generate docking poses (one 400-generation run per FF configuration, per complex).Figure 4. Strategy used in the force field parameter tuning protocol. ‘Decoy poses’ are non-native-like, to be distinguished from the native-like by a proper choice of FF parameters. These three parameters were subjected to a systematic scan. Within the scanned parameter space volume, the representative subset of possible FF configurations includes: Core FF, Preliminary FF setups, setups at the extremes (corners) and at the center of the scanned parameter ‘cube’. This representative subset is used to generate docking poses (one 400-generation run per FF configuration, per complex).
- Last, a systematic scan of hydrophobicity parameters associated to considered classes of carbons was performed. The 33 carbon force field types were regrouped into 3 different hydrophobicity classes: aliphatic, aromatic and polarized, each being attributed a hydrophobicity constant Kc (see Table 1). A total of 10 discrete values (0; 0.010; 0.025; 0.050; 0.075; 0.100; 0.150; 0.200; 0.250; 0.300) are considered as possible Kc choices, allowing for 1000 combinations to scan. The scan was bound to highlight a triplet which systematically ranks, for a large majority of Astex/CCDC-clean complexes, native-like poses as lowest-energy states, out of large and decoy-rich sets of poses. Pose sets were generated by S4MPLE, in iteratively repeated runs employing various FF parameterization schemes (see Figure 4). In practice and for each complex, the pool of poses is rescored using the fitted parameters and one of the 1000 combinations. The top-ranked pose is saved for each complex and all tested combinations, thus it is possible to extract triplets which most frequently favor the expected binding mode. Validation of the herein obtained Fit FF configuration was done by docking of Astex Diverse ligands, and assessment of redocking success.
2.8. Redocking Protocol Using S4MPLE
- computing partial charges (Gasteiger type) using ChemAxon libraries [59]
- generating a single conformer using ChemAxon libraries (avoiding to start from the expected solution, when the docking accuracy may be artificially enhanced)
2.9. Redocking Protocol Using FlexX
3. Results and Discussion
3.1. Force Field Fitting
3.1.1. Tuning of Desolvation Contributions
3.1.2. Optimizing Hydrophobic Contact Strengths
| Number of Successfully Predicted Complexes | Weights (K) | ||
|---|---|---|---|
| Kpolarized | Karom | Kaliph | |
| 154/191 | 0.000 | 0.075 | 0.150 |
| 0.000 | 0.100 | 0.150 | |
| 0.000 | 0.150 | 0.150 | |
| 0.000 | 0.150 | 0.200 | |
| 0.000 | 0.200 | 0.200 | |
| 0.010 | 0.075 | 0.150 | |
| 0.010 | 0.100 | 0.150 | |
| 0.010 | 0.150 | 0.150 | |
| 0.010 | 0.200 | 0.200 | |
| 0.025 | 0.150 | 0.150 | |
| 0.200 | 0.150 | 0.150 | |
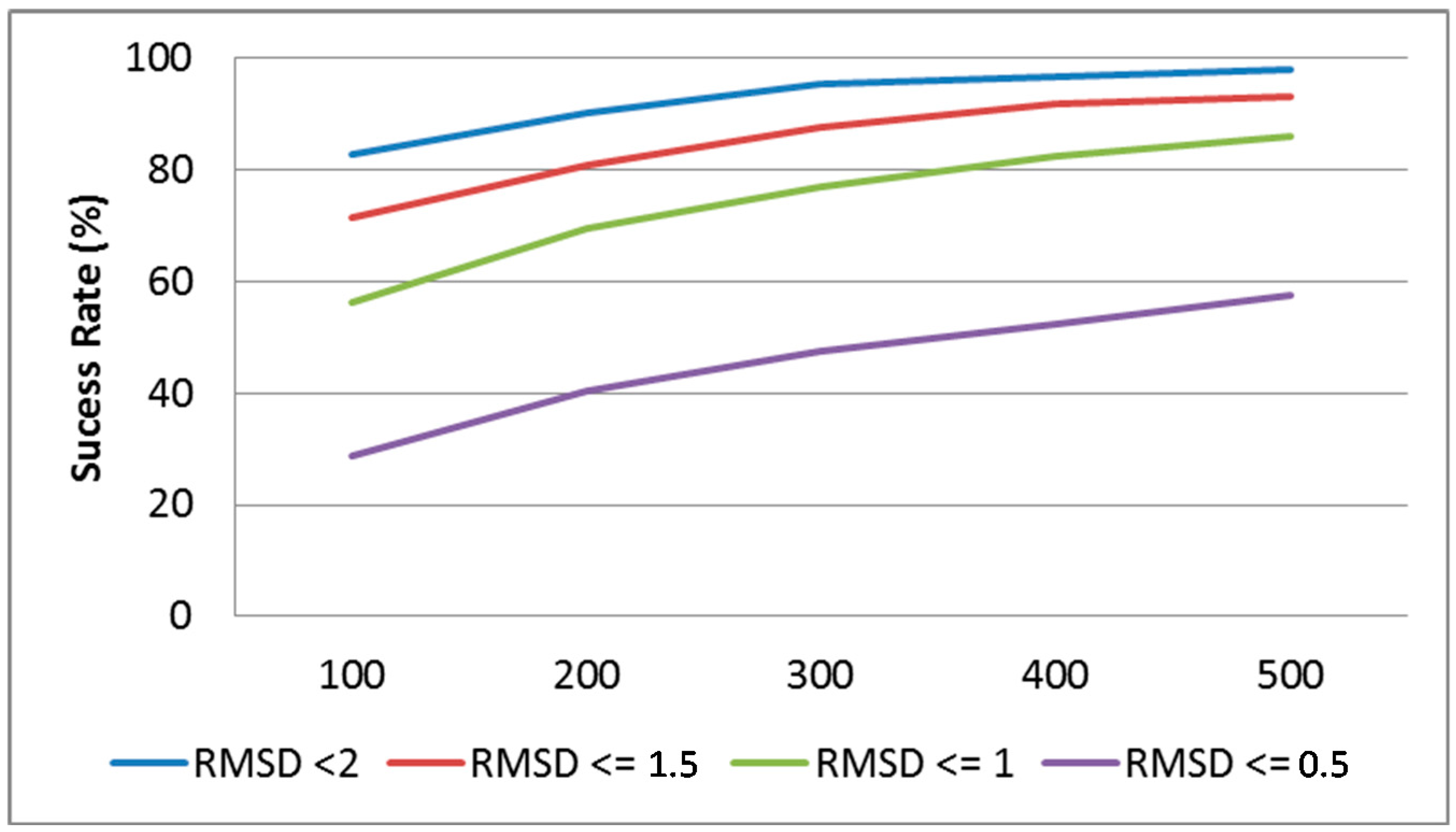
3.2. Redocking-Astex Diverse Set
| Docking Tools (Scoring) | Success Rate (%) Top Ranked-Pose | Success Rate (%) Saved Poses |
|---|---|---|
| S4MPLE (Core FF) | 76 | 93 |
| S4MPLE (Fit FF) | 85 | 96 |
| FlexX * | 71 | 91 |
| GOLD (Goldscore) ** [19] | 75–81 | Unavailable |
| Plants (ChemPLP) [65] | 87 | 97 |
| Plants (PLP) [65] | 84 | |
| LGA (LargeAll) [34] | 63 | Unavailable |
| RosettaLig [66] | 58 | 92 |
| SKATE [67] | 87 | 98 |

3.2.1. Insights on Failure Cases
- All-atom RMSD is high, albeit the top-ranked pose is chemically meaningful, reproducing all the key ligand-site contacts. This may happen:
- (a)
- if the ligand includes a moiety dangling out in the solvent, or
- (b)
- the ligand is ‘pseudo-symmetric’, in the sense that it features chemically similar groups which could, in principle, compete for a same binding spot.
- 2.
- Neither the top-ranked pose, nor any other of sampled poses, match the native one. This may be due to:
- (a)
- insufficient sampling,
- (b)
- inappropriate definition of the binding site (in redocking, this includes ignoring key waters mediating ligand-site interactions), or
- (c)
- a highly unrealistic force field setup, causing native-like structures to be of high energy, thus effectively preventing them from being sampled and saved.
- 3.
- The top-ranked pose differs from the native, but the latter is being sampled and listed among less stable ones. This may be:
- (a)
- imputable to the force field setup, counting as a failure to identify the expected binding mode as the absolute energy minimum, or
- (b)
- an inappropriate definition of the binding site (like in 2(b)—however, of more limited impact on results since the expected binding mode has been saved), or
- (c)
- an entropic effect: minima deeper than the native one exist, as predicted by the FF, but are narrow and therefore not significantly populated at room temperature.
- 1G9V, 1GM8 & 1HVY ligands have limited direct contacts with the binding site: most of the ligand/site hydrogen bonds are bridged with waters. Worse, these ligands feature solvated carboxylate groups, not directly interacting with the target. In 1HVY, the dihydroquinazoline moiety from drug Tomudex is perfectly docked, whereas the two loose carboxylates are, in absence of crystallographic waters, attracted towards neighboring basic residues. Therefore, direct site-ligand contacts are all well predicted—this fits scenario (1(a)). Same holds for 1GM8, the hydrophobic phenylacetamide moiety is correctly positioned, unlike the anionic -lactam moiety. 1G9V, however, is totally dependent on specific water-mediated interactions, therefore classifying as failure 3(b) (the correct pose is being sampled, but not top ranked).
- 2BR1 & 1XM6 are further examples of (3(b)) complexes where water-mediated interactions play an important role. By contrast to the examples above, these water-mediated contacts occur deeply within the binding site. In the top-ranked pose of 2BR1, the ligand is bound head-to-tail, in burying the methoxyphenyl moiety into a deep pocket filled with water in the X-ray structure. In 1XM6, the location of the propoxyphenyl group is perfect but the oxazolidone moiety is erroneously predicted to directly interact with a Zn2+ ion (in the experimental structure, a water molecule occupies this area).
- 1GPK & 1MEH are classical FF failure cases (3(a)), native conformations being sampled but not top-ranked. The best poses favor an ionic bond instead of two hydrogen bonds, observed in the PDB file, showing that fine tuning of ionic/polar interactions and desolvation penalty is not perfect. In 1GPK the top-ranked pose is inverted, but nevertheless fulfills expected hydrophobic contacts with the site. In 1MEH, the solvent-exposed carboxylate unsurprisingly prefers the neighborhood of R414, instead of the crystallographic hydrogen bonds to S262. This has a very limited impact on the correctly predicted pose of the buried aromatic moiety.
- 1HP0 is an interesting case, where a non-native conformer with flipped deazaadenine group is nevertheless almost correctly docked—fulfilling all of the experimentally observed contacts. The ligand adopts a correct binding mode around the calcium ion, and reproduces the stacking interactions of the flipped aromatic moiety. Non-bonded energies are similar, thus the preference for the native conformer should have been played out in terms of intra-ligand strain contributions. This is not happening (the native conformer is visited, but ranked slightly less well). At this point, it is difficult to formally rank this as a force field failure 3(a), rather than a genuine multiple binding mode example 1.b. 1JJE is in a similar situation, featuring a pseudo-symmetric ligand, derivative of 2,3-dibenzylsuccinic acid in which one of the Phe groups ports a -O-CH2-O- bridge (benzodioxole). The latter makes, however, no additional strong contact to the protein, whereas the central, succinyl moiety coordinating both Zn2+ ions is perfectly predicted. It seems that the benzodioxole group may be equally well accommodated on each side of the binding site. Docking does not favor the one expected. It is not clear, however, whether the experimentally observed electron density is witness of a single binding mode, or whether it is a statistical expression of a slight preference among two almost equally populated modes. The head-to-tail binding of this pseudo-symmetric ligand results in an impressive RMSD of 8 Å.
- In 1TZ8, the native PDB pose surprisingly exhibits several slight ligand-site clashes (e.g., distance L17_CD1-Ligand_C = 3.0 Å). Expectedly, even the Fit FF with slightly downscaled repulsive vdW terms, leads to a wrong top-ranked pose—albeit it enumerates the native one. It should be noted that this complex is often reported as a failure [66,67]. Furthermore, authors of the Astex compilation dataset reported this complex as borderline in term of quality of the ligand’s electronic density [55]. Hence, this case should be most likely classified as 3(b) rather than a FF problem (3(a)).
- 1YGC: the most solvent-exposed moiety (arylsulphonamide) adopts a wrong alternative conformation (direct hydrogen-bond with the C58 backbone instead of two water-mediated hydrogen bonds with Y94 and T98), whereas the rest of this large ligand is properly docked.
3.2.2. FF-Based Scoring: Core vs. Fit
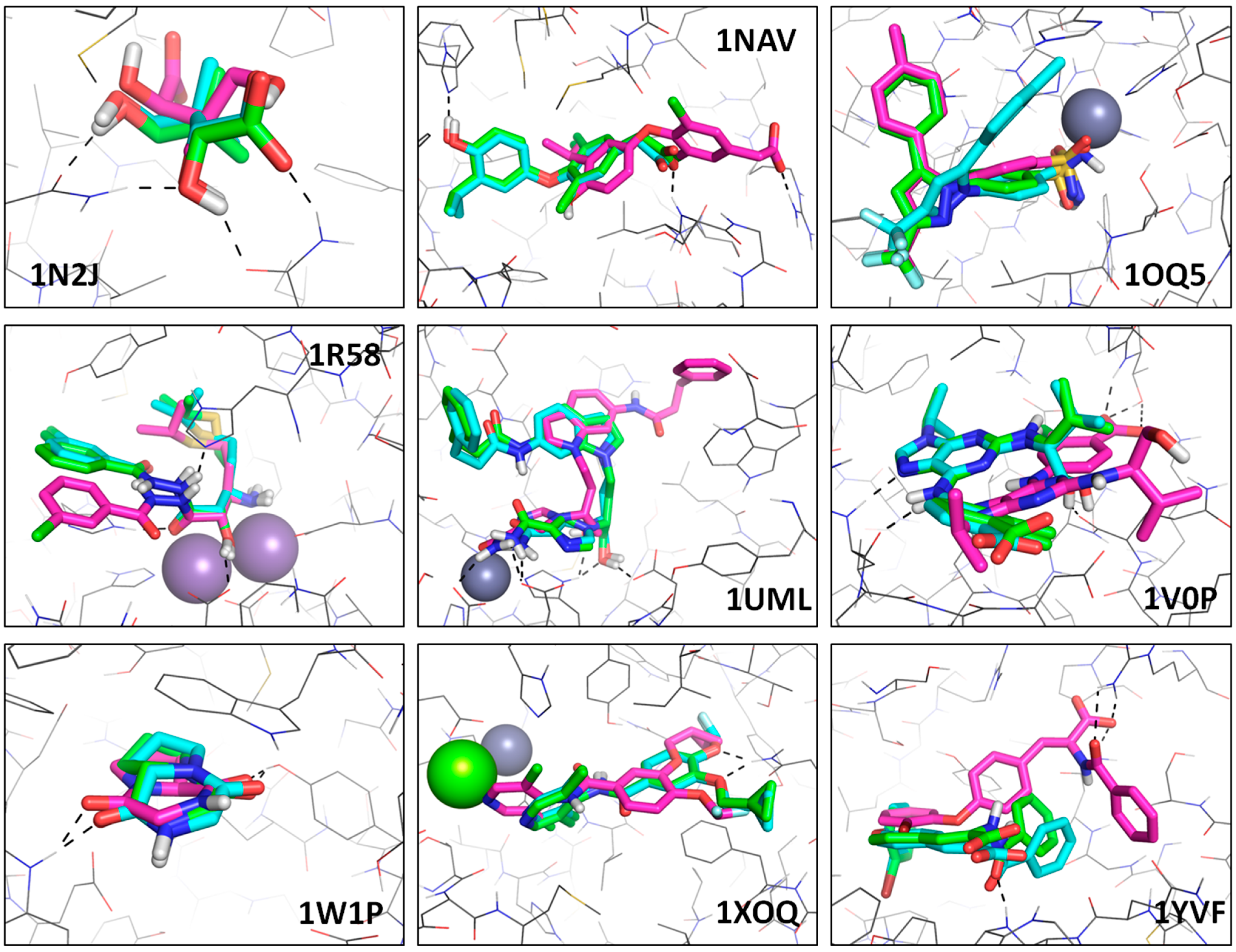
| PDB | Core FF | Fit FF |
|---|---|---|
| 1N2J | 3.85 | 1.42 |
| 1NAV | 6.26 | 0.35 |
| 1OQ5 | 0.92 | 2.83 |
| 1R58 | 3.03 | 0.74 |
| 1UML | 7.59 | 0.66 |
| 1V0P | 7.60 | 0.41 |
| 1W1P | 2.94 | 0.39 |
| 1XOQ | 4.04 | 0.30 |
| 1YVF | 6.11 | 0.89 |
- 1OQ5: Fit FF has the second-ranked pose matching the expected binding mode, but the most stable one appears clearly non-native whereas Core FF selects the native one. In this Fit FF failure, the ligand is rotated around the pyrazole-benzenesulphonamide axis, with the tolyl group occupying an alternative sub-pocket.
- 1N2J: this small ligand (pantoate) makes four direct hydrogen bonds with the binding site. With Core FF, the top-ranked pose does not make any of these, but places the carboxylate in the hydrophobic area where the two -Me groups are expected. Favorable contact terms (hydrogen-bond and hydrophobic enclosure), and desolvation (penalizing the burial of a -COO− in a hydrophobic pocket) in Fit FF successfully restore the correct binding mode at the top of the list.
- 1NAV: the compound is buried and makes several hydrogen bonds with the binding site. Fit FF finds the native binding mode with a great precision (RMSD ≈ 0.5 Å), whereas Core FF selects a translated pose (several Å away from the experimental binding mode: RMSD ≈ 6 Å) in order to create an ionic bond between the carboxylic group of the ligand and the solvent-exposed R266. This is another example of overestimated polar contributions in absence of scaled down electrostatics and desolvation.
- 1R58: in that case, both scoring schemes lead to an acceptable solution for buried and metal coordination groups of the ligand, but the chlorophenyl moiety is wrongly docked by Core FF, which has no particular incentive to flip this group in order to bring in within hydrophobic contact range with Y444, as seen in the experimental structure.
- 1UML: this ligand is located near a Zn2+ ion but does not directly interact with it in the X-ray structure. Fit FF leads to a perfect pose, while the Core FF completely misplaces the ligand, in order to allow for an interaction with that cation.
- 1V0P: Fit FF returns a perfect top-ranked solution, in which the carboxylate group of the ligand is solvent-exposed and doesn’t make any direct contact with the protein. At the opposite, the Core FF favors a pose where the ligand is less buried, allowing the carboxylate group to make several fake hydrogen bonds with the binding site.
- 1W1P: this complex contains a small cyclic dipeptide (Gly-L/Pro). Although both hydrogen bonds with the site are conserved using the Core FF, the RMSD is high because the ligand is flipped with respect to the experimental binding mode. This is rather a 1.a scenario, in which ligand flipping does not affect observed interaction patterns. Conversely, the Fit FF top-ranked pose matches the exact binding mode.
- 1XOQ: Although two cations are present in the binding site (Zn2+ and Mg2+), the drug (roflumilast) does not directly interact with them in the crystal structure. The Core FF forces a binding mode where the dichloropyridine group of the ligand is close to the magnesium ion, whereas the Fit FF returns the expected binding mode with two hydrogen-bonds with the same sidechain (Q369).
- 1YVF: this case is similar to 1V0P, featuring a solvent-exposed carboxylate, forced by Core FF into several non-native interactions with R394. The best pose from the Fit FF reproduces almost exactly the experimental binding mode. Generally speaking, Core FF favors less buried ligand poses, with fake polar contacts: it overestimates Coulomb terms, ignores desolvation and does not reward hydrophobic contacts.
4. Conclusions
- (a)
- a plethora of technical but important parameters—such as, for example, stopping criteria of local search procedures, etc.—were not yet optimized, being set by default to rather strict values.
- (b)
- the main goal behind this development is not to add one more rigid-site docking program to an already long list. So far, the purpose of this development was to assess in how far experimental docking poses can be correctly predicted on the basis of the herein defined energy function (no re-ranking based on fitted free energy scoring functions) if unbiased sampling is allowed.
Supplementary Material
- x86_64 executable of S4MPLE,
- User guide,
- various ligand preparation tools and
- force field parameter distributions
- spreadsheet with the list of the training set compounds used to calibrate Fit FF, and detailed docking results for each of the repeated docking runs are available for download from: http://infochim.u-strasbg.fr, DOWNLOADS section (tar.gz). Docked poses are available upon request (mol2 format) (http://creativecommons.org/licenses/by/4.0/).
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Michel, J.; Foloppe, N.; Essex, J.W. Rigorous free energy calculations in structure-based drug design. Mol. Inf. 2010, 29, 570–578. [Google Scholar] [CrossRef]
- Sperandio, O.; Miteva, M.A.; Villoutreix, B.O. Combining ligand- and structure-based methods in drug design projects. Curr. Comput. -Aid. Drug 2008, 4, 250–258. [Google Scholar] [CrossRef]
- Blundell, T.L. Structure-based drug design. Nature 1996, 384, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Abrams, C.F.; Vanden-Eijnden, E. Large-scale conformational sampling of proteins using temperature-accelerated molecular dynamics. Proc. Natl. Acad. Sci. USA 2010, 107, 4961–4966. [Google Scholar] [CrossRef] [PubMed]
- Foloppe, N.; Chen, I.J. Conformational sampling and energetics of drug-like molecules. Curr. Med. Chem. 2009, 16, 3381–3413. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.C.; Wolohan, P.R.N.; Abrahamian, E.; Clark, R.D. Parameterization and conformational sampling effects in pharmacophore multiplet searching. J. Chem. Inf. Model. 2008, 48, 2326–2334. [Google Scholar] [CrossRef] [PubMed]
- Parenti, M.D.; Rastelli, G. Advances and applications of binding affinity prediction methods in drug discovery. Biotechnol. Adv. 2012, 30, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.C.; Xiong, B.; Zheng, M.Y.; Luo, X.M.; Luo, C.; Liu, X.A.; Du, Y.; Li, J.; Zhu, W.L.; Shen, J.K.; et al. Knowledge-based scoring functions in drug design: 2. Can the knowledge base be enriched? J. Chem. Inf. Model. 2011, 51, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Von Korff, M.; Freyss, J.; Sander, T. Comparison of Ligand- and Structure-Based Virtual Screening on the Dud Data Set. In Proceedings of the 8th International Conference on Chemical Structures, Noordwijkerhout, The Netherlands, 1–5 June 2008; Amer Chemical Soc: Noordwijkerhout, The Netherlands; pp. 209–231.
- Tantar, A.-A.; Conilleau, S.; Parent, B.; Melab, N.; Brillet, L.; Roy, S.; Talbi, E.-G.; Horvath, D. Docking and biomolecular simulations on computer grids: Status and trends. Curr. Comput. -Aid. Drug 2008, 4, 235–249. [Google Scholar] [CrossRef]
- McGann, M. Fred pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Ewing, T.J.; Makino, S.; Skillman, A.G.; Kuntz, I.D. Dock 4.0: Search strategies for automated molecular docking of flexible molecule databases. J. Comput. Aided Mol. Des. 2001, 15, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Claussen, H.; Buning, C.; Rarey, M.; Lengauer, T. Flexe: Efficient molecular docking considering protein structure variations. J. Mol. Biol. 2001, 308, 377–395. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of autodock. J. Mol. Recognit. 1996, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Huey, R.; Olson, A.J. Distributed automated docking of flexible ligands to proteins: Parallel applications of autodock 2.4. J. Comput. Aided Mol. Des. 1996, 10, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.; Leach, A.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using gold. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Meiler, J.; Baker, D. Rosettaligand: Protein-small molecule docking with full side-chain flexibility. Proteins 2006, 65, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sanner, M.F. Flipdock: Docking flexible ligands into flexible receptors. Proteins 2007, 68, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Moitessier, N. Docking ligands into flexible and solvated macromolecules. 3. Impact of input ligand conformation, protein flexibility, and water molecules on the accuracy of docking programs. J. Chem. Inf. Model. 2009, 49, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Englebienne, P.; Yannopoulos, C.G.; Chan, L.; Das, S.K.; Bilimoria, D.; L’Heureux, L.; Moitessier, N. Docking ligands into flexible and solvated macromolecules. 2. Development and application of fitted 1.5 to the virtual screening of potential HCV polymerase inhibitors. J. Chem. Inf. Model. 2008, 48, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Englebienne, P.; Moitessier, N. Docking ligands into flexible and solvated macromolecules. 1. Development and validation of fitted 1.0. J. Chem. Inf. Model. 2007, 47, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Sinko, W.; de Oliveira, C.A.F.; Pierce, L.C.T.; McCammon, J.A. Protecting high energy barriers: A new equation to regulate boost energy in accelerated molecular dynamics simulations. J. Chem. Theor. Comput. 2012, 8, 17–23. [Google Scholar] [CrossRef]
- Omelyan, I.P.; Kovalenko, A. Overcoming the barrier on time step size in multiscale molecular dynamics simulation of molecular liquids. J. Chem. Theor. Comput. 2012, 8, 6–16. [Google Scholar] [CrossRef]
- Hsu, H.-P.; Grassberger, P. A review of monte carlo simulations of polymers with perm. J. Stat. Phys. 2011, 144, 597–637. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. Eadock: Docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins 2007, 67, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Schefzick, S.; Bradley, M. Comparison of commercially available genetic algorithms: Gas as variable selection tool. J. Comput. Aided Mol. Des. 2004, 18, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R. Flexible ligand docking using evolutionary algorithms: Investigating the effects of variation operators and local search hybrids. Biosystems 2003, 72, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Grosdidier, A.; Cuendet, M.; Michielin, O. Use of the facts solvation model for protein-ligand docking calculations. Application to eadock. J. Mol. Recognit. 23, 457–461. [CrossRef]
- Fuhrmann, J.; Rurainski, A.; Lenhof, H.P.; Neumann, D. A new lamarckian genetic algorithm for flexible ligand-receptor docking. J. Comput. Chem. 2010, 31, 1911–1918. [Google Scholar] [PubMed]
- Neudert, G.; Klebe, G. Dsx: A knowledge-based scoring function for the assessment of protein-ligand complexes. J. Chem. Inf. Model. 2011, 51, 2731–2745. [Google Scholar] [CrossRef] [PubMed]
- Bissantz, C.; Folkers, G.; Rognan, D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 2000, 43, 4759–4767. [Google Scholar] [CrossRef] [PubMed]
- Hoffer, L.; Renaud, J.-P.; Horvath, D. In silico fragment-based drug discovery: Setup and validation of a fragment-to-lead computational protocol using S4MPLE. J. Chem. Inf. Model. 2013, 53, 836–851. [Google Scholar] [CrossRef] [PubMed]
- Hoffer, L.; Horvath, D. S4MPLE—Sampler for multiple protein-ligand entities: Simultaneous docking of several entities. J. Chem. Inf. Model. 2012, 53, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Darden, T.A.; Cheatham, T.E.; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham, T.E.; Debolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. Amber a package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular-dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Wang, J.M.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Halperin, I.; Ma, B.; Wolfson, H.; Nussinov, R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins 2002, 47, 409–443. [Google Scholar] [CrossRef] [PubMed]
- Parent, B.; Kökösy, A.; Horvath, D. Optimized evolutionary strategies in conformational sampling. Soft Comput. 2007, 11, 63–79. [Google Scholar] [CrossRef]
- Marcou, G.; Rognan, D. Optimizing fragment and scaffold docking by use of molecular interaction fingerprints. J. Chem. Inf. Model. 2007, 47, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Brewerton, S.C. The use of protein-ligand interaction fingerprints in docking. Curr. Opin. Drug Discov. Devel. 2008, 11, 356–364. [Google Scholar] [PubMed]
- Deng, Z.; Chuaqui, C.; Singh, J. Structural interaction fingerprint (sift): A novel method for analyzing three-dimensional protein-ligand binding interactions. J. Med. Chem. 2004, 47, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Dauberosguthorpe, P.; Roberts, V.A.; Osguthorpe, D.J.; Wolff, J.; Genest, M.; Hagler, A.T. Structure and energetics of ligand-binding to proteins: Escherichia-coli dihydrofolate reductase trimethoprim, a drug-receptor system. Proteins 1988, 4, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Tantar, A.-A.; Melab, N.; Talbi, E.-G.; Parent, B.; Horvath, D. A parallel hybrid genetic algorithm for protein structure prediction on the computational grid. Future Gen. Comput. Syst. 2007, 23, 398–409. [Google Scholar] [CrossRef]
- Halgren, T.A. MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries. J. Comput. Chem. 1999, 20, 730–748. [Google Scholar] [CrossRef]
- Tripos, I. Sybyl, version 8.0; Tripos: St. Louis, MO, USA, 2007.
- Horvath, D. A virtual screening approach applied to the search for trypanothione reductase inhibitors. J. Med. Chem. 1997, 40, 2412–2423. [Google Scholar] [CrossRef] [PubMed]
- Mazur, J.; Jernigan, R. Distance-dependent dielectric-constants and their application to double-helical DNA. Biopolymers 1991, 31, 1615–1629. [Google Scholar] [CrossRef] [PubMed]
- Nissink, J.W.; Murray, C.; Hartshorn, M.; Verdonk, M.L.; Cole, J.C.; Taylor, R. A new test set for validating predictions of protein-ligand interaction. Proteins 2002, 49, 457–471. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The Pymol Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Hartshorn, M.J.; Verdonk, M.L.; Chessari, G.; Brewerton, S.C.; Mooij, W.T.; Mortenson, P.N.; Murray, C.W. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007, 50, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Rcsb Protein Data Bank. Available online: http://www.rcsb.org/pdb/ (accessed on 18 May 2015).
- Horvath, D.; Brillet, L.; Roy, S.; Conilleau, S.; Tantar, A.-A.; Boisson, J.-C.; Melab, N.; Talbi, E.-G. Local vs. Global search strategies in evolutionary grid-based conformational sampling & docking. In Proceedings of the IEEE Congress on Evolutionary Computation CEC 09, IEEE, Trondheim, Norway, 18–21 May 2009; pp. 247–254.
- Parent, B.; Tantar, A.; Melab, N.; Talbi, E.-G.; Horvath, D. Grid-Based Evolutionary Strategies Applied to the Conformational Sampling Problem. In Proceedings of the IEEE Congress on Evolutionary Computation, CEC 2007, Singapore, 25–28 September 2007; pp. 291–296.
- ChemAxon. Calculation of Partial Charge Distributions. Available online: http://www.chemaxon.com/marvin/help/calculations/charge.html (accessed on 15 February 2009).
- Wang, J.M.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- MOE (Molecular Operating Environment), version 2005.06; Chemical Computing Group, Inc.: Montreal, QC, Canada, 2005.
- Kellenberger, E.; Rodrigo, J.; Muller, P.; Rognan, D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 2004, 57, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Kramer, B.; Rarey, M.; Lengauer, T. Evaluation of the flexx incremental construction algorithm for protein-ligand docking. Proteins 1999, 37, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Horvath, D.; Lippens, G.; vanBelle, D. Development and parametrization of continuum solvent models. 2. A unified approach to the solvation problem. J. Chem. Phys. 1996, 105, 4197–4210. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with plants. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.W.; Baker, D. Rosettaligand docking with full ligand and receptor flexibility. J. Mol. Biol. 2009, 385, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.A.; Marshall, G.R. Skate: A docking program that decouples systematic sampling from scoring. J. Comput. Chem. 2010, 31, 2540–2554. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffer, L.; Chira, C.; Marcou, G.; Varnek, A.; Horvath, D. S4MPLE—Sampler for Multiple Protein-Ligand Entities: Methodology and Rigid-Site Docking Benchmarking. Molecules 2015, 20, 8997-9028. https://doi.org/10.3390/molecules20058997
Hoffer L, Chira C, Marcou G, Varnek A, Horvath D. S4MPLE—Sampler for Multiple Protein-Ligand Entities: Methodology and Rigid-Site Docking Benchmarking. Molecules. 2015; 20(5):8997-9028. https://doi.org/10.3390/molecules20058997
Chicago/Turabian StyleHoffer, Laurent, Camelia Chira, Gilles Marcou, Alexandre Varnek, and Dragos Horvath. 2015. "S4MPLE—Sampler for Multiple Protein-Ligand Entities: Methodology and Rigid-Site Docking Benchmarking" Molecules 20, no. 5: 8997-9028. https://doi.org/10.3390/molecules20058997