Genetic Authentication of Gardenia jasminoides Ellis var. grandiflora Nakai by Improved RAPD-Derived DNA Markers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Cloning of RAPD Amplification Fragments
| Sample | Species | Source | No. |
|---|---|---|---|
| JX | G. jasminoides Ellis var. grandiflora Nakai | Shangrao, Jiangxi | 1 |
| FJ | G. jasminoides Ellis var. grandiflora Nakai | Fuzhou, Fujian | 2 |
| GZ | G. jasminoides Ellis var. grandiflora Nakai | Anshun, Guizhou | 3 |
| SC | G. jasminoides Ellis var. grandiflora Nakai | Luzhou, Sichuan | 4 |
| HN | G. jasminoides Ellis var. grandiflora Nakai | Changsha, Hunan | 5 |
| ZJ | G. jasminoides Ellis var. grandiflora Nakai | Lishui, Zhejiang | 6 |
| SC2 | G. jasminoides (G. jasminoides J. Ellis) | Luzhou, Sichuan | 7 |
| GD | G. jasminoides Ellis var. grandiflora Nakai | Shenzhen, Guangdong | 8 |
| FJ2 | G. jasminoides Ellis var. grandiflora Nakai | Quanzhou, Fujian | 9 |
| HN2 | G. jasminoides Ellis var. grandiflora Nakai | Changsha, Hunan | 10 |
| AH | G. jasminoides Ellis var. grandiflora Nakai | Huainan, Aihui | 11 |
| JX2 | G. jasminoides (G. jasminoides J. Ellis) | Fuzhou, Jiangxi | 12 |
| JS | G. jasminoides (G. jasminoides J. Ellis) | Nanjing, Jiangsu | 13 |
| HN3 | G. jasminoides (G. jasminoides J. Ellis) | Changsha, Hunan | 14 |
| SX | G. jasminoides (G. jasminoides J. Ellis) | Taiyuan, Shanxi | 15 |
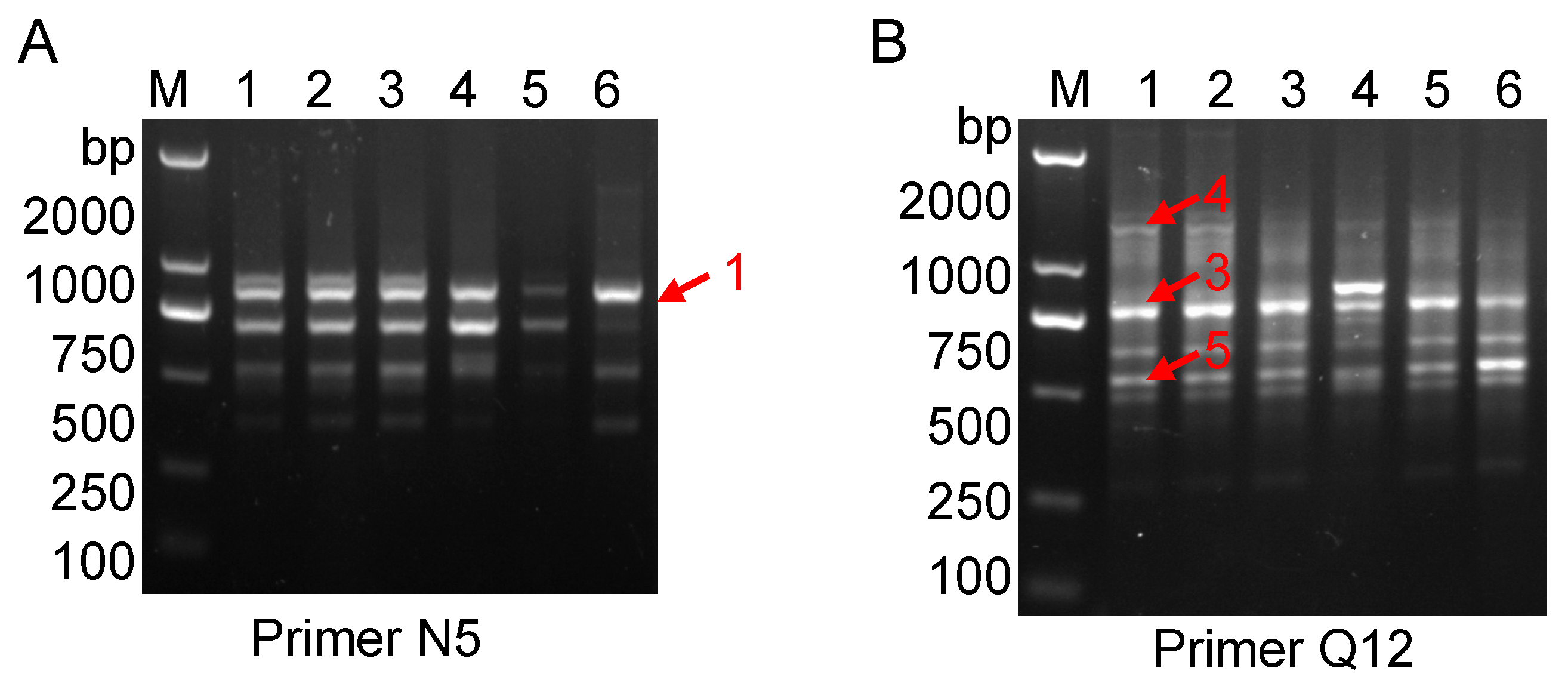

2.1.2. Sequencing and Characterization of G. jasminoides Ellis var. grandiflora Nakai Specific RAPD Fragments
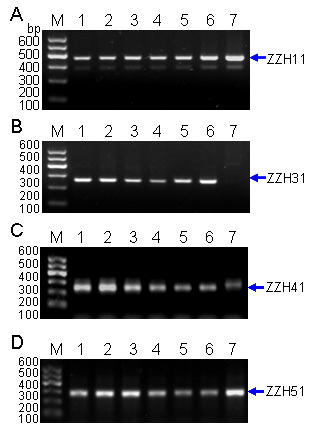
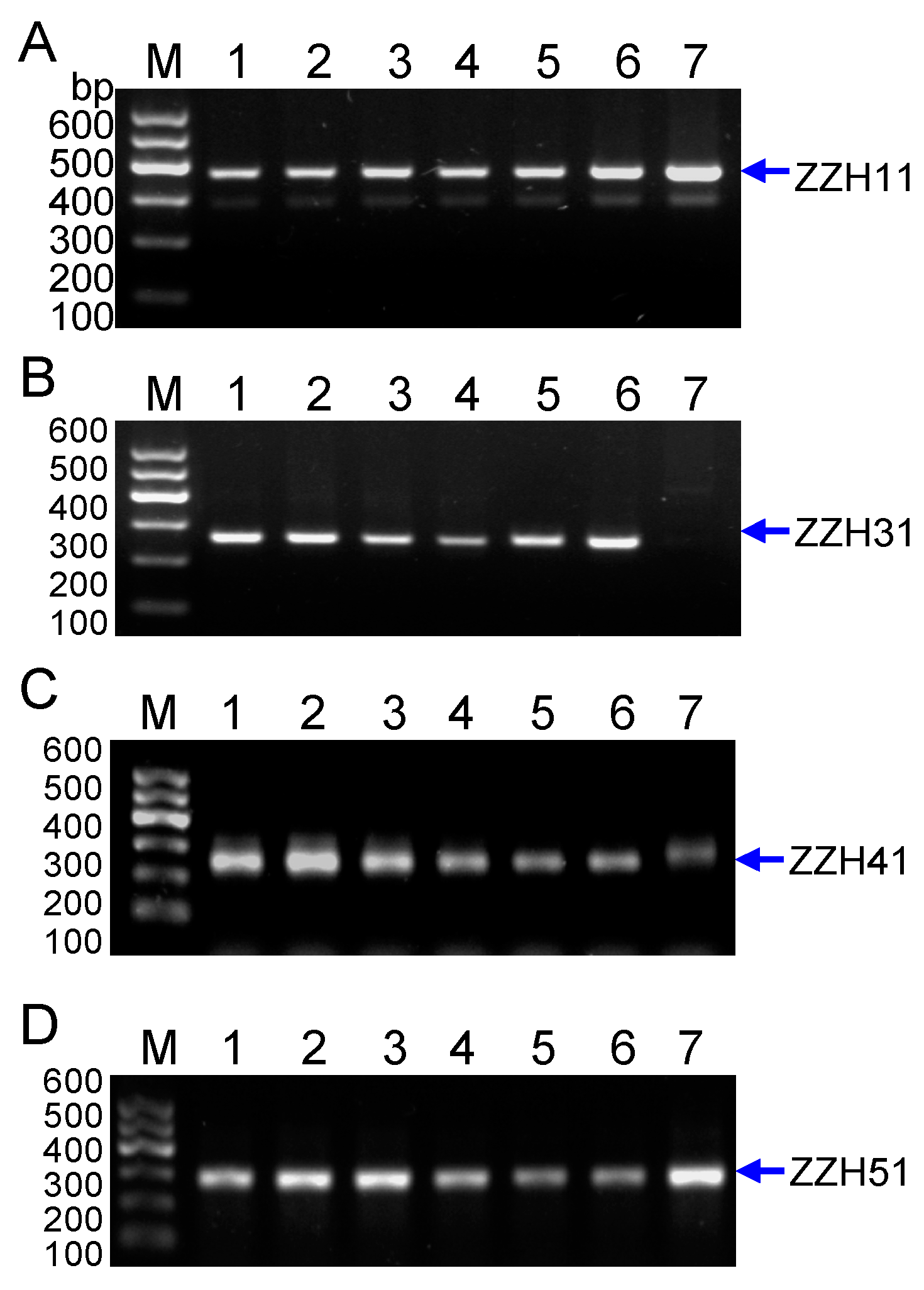
2.1.3. Development of Specific SCAR Markers for G. jasminoides Ellis var. grandiflora Nakai, and Analysis of the PCR Amplicons at Different Species

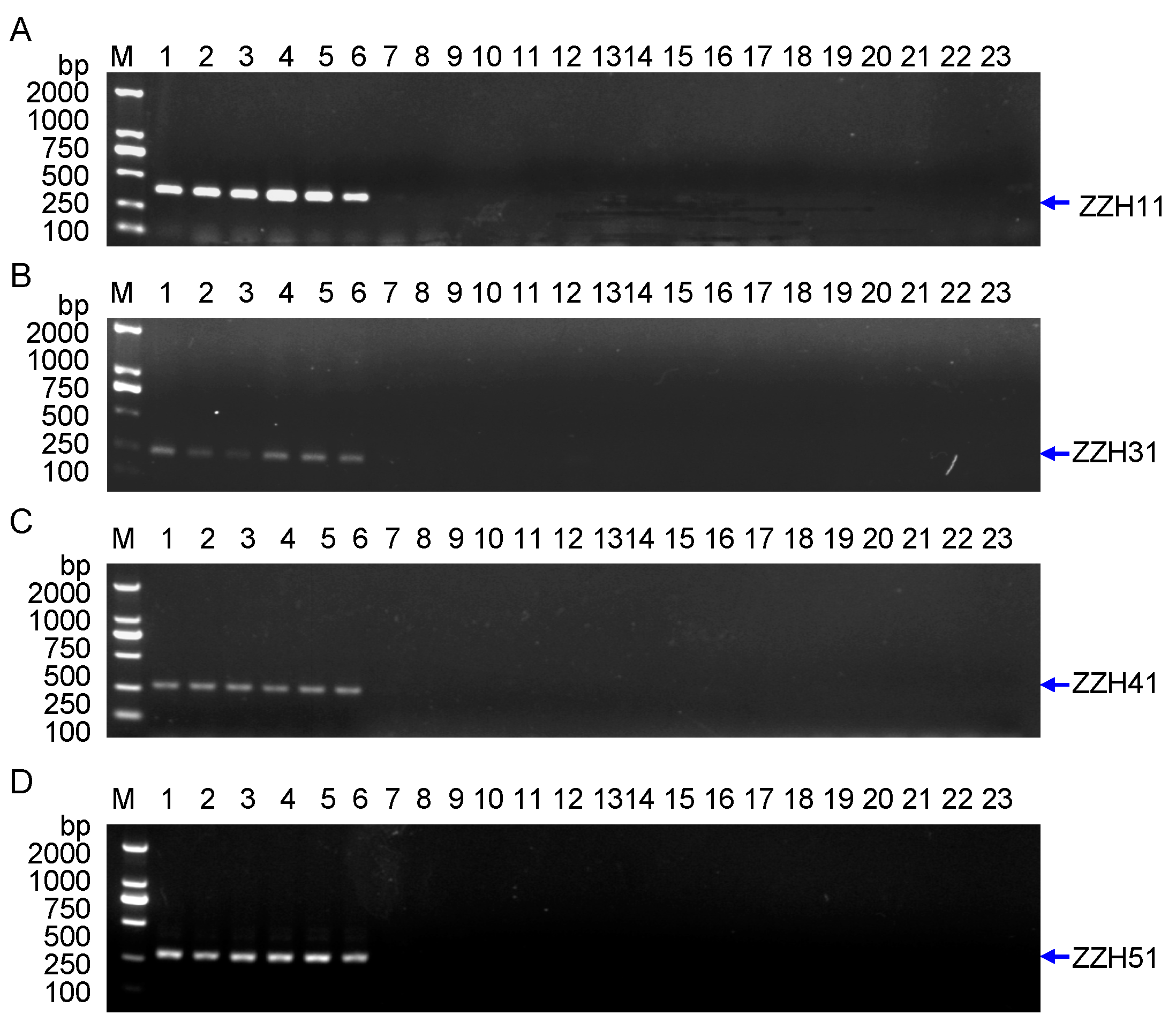
| SCAR | 5′-primer | Sequence (5′–3′) | 3′-primer | Sequence (5′–3′) | Size | Tm (°C) |
|---|---|---|---|---|---|---|
| ZZH11 | ZZH11-L | TGCCAATGATCGAGGTTCTC | ZZH11-R | GCACTACAACCACCCAAGGA | 381 | 60 |
| ZZH31 | ZZH31-L | TTCCAAACCACAACAAGCCGT | ZZH31-R | GCATGAGTTTGTCGGCGATTT | 245 | 64 |
| ZZH41 | ZZH41-L | ATGGCGTACCTTCCAATGTG | ZZH41-R | AATGGCCAGACCATGTCAAG | 279 | 60 |
| ZZH51 | ZZH51-L | ACGCTAAGGAGGATCACGACA | ZZH51-R | ACTGACAAGACGGACTGCAAC | 291 | 60 |
2.2. Discussion

3. Experimental Section
3.1. DNA Extraction from G. jasminoides
3.2. Amplification of DNA by Improved RAPD
3.3. Cloning, Identification and Sequencing of DNA Fragments
3.4. Bioinformatic Analysis by Online Program BLAST
3.5. Design of SCAR Primers
3.6. Development SCAR Markers and SCAR Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Choi, S.J.; Kim, M.J.; Heo, H.J.; Hong, B.; Cho, H.Y.; Kim, Y.J.; Kim, H.K.; Lim, S.T.; Jun, W.J.; Kim, E.K.; et al. Ameliorating effect of Gardenia jasminoides extract on amyloid beta peptide-induced neuronal cell deficit. Mol. Cells 2007, 24, 113–118. [Google Scholar] [PubMed]
- Chen, Y.H.; Lan, T.; Li, J.; Qiu, C.H.; Wu, T.; Gou, H.J.; Lu, M.Q. Gardenia jasminoides attenuates hepatocellular injury and fibrosis in bile duct-ligated rats and human hepatic stellate cells. World J. Gastroenterol. 2012, 18, 7158–7165. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Jeong, C.S.; Moon, A. Genipin, a constituent of Gardenia jasminoides Ellis, induces apoptosis and inhibits invasion in MDA-MB-231 breast cancer cells. Oncol. Rep. 2012, 27, 562–572. [Google Scholar]
- Yang, Q.; Wu, B.; Shi, Y.; Du, X.; Fan, M.; Sun, Z.; Cui, X.; Huang, C. Guided fractionation and analysis of compounds with anti-influenza virus activity from Gardenia jasminoides Ellis. Arch. Pharm. Res. 2012, 35, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Xiaofeng, Y.; Qinren, C.; Jingping, H.; Xiao, C.; Miaomiao, W.; Xiangru, F.; Xianxing, X.; Meixia, H.; Jing, L.; Jingyuan, W.; et al. Geniposide, an iridoid glucoside derived from Gardenia jasminoides, protects against lipopolysaccharide-induced acute lung injury in mice. Planta Med. 2012, 78, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, J.; Xi, L.; Qian, Z.; Wang, Z.; Yang, L. Protective effect of crocetin on hemorrhagic shock-induced acute renal failure in rats. Shock 2012, 38, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, F.; Shimazawa, M.; Umigai, N.; Ogishima, H.; Nakamura, S.; Tsuruma, K.; Hara, H. Crocetin, a carotenoid derivative, inhibits retinal ischemic damage in mice. Eur. J. Pharmacol. 2013, 703, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Lao, W.; Xiao, L.; Wang, Z.; Kamal, M.A.; Seale, J.P.; Qu, X. Managing the combination of nonalcoholic fatty liver disease and metabolic syndrome with Chinese herbal extracts in high-fat-diet fed rats. Evid. Based Complement. Altern. Med. 2013, 2013, 306738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Liu, H.; Yang, M.; Wei, S.F. Antithrombotic activi ueous extract from Gardenia jasminoides and its main constituent. Pharm. Biol. 2013, 51, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.Y.; Lee, A.Y.; Kim, H.K. The Gardenia jasminoides extract and its constituent, geniposide, elicit anti-allergic effects on atopic dermatitis by inhibiting histamine in vitro and in vivo. J. Ethnopharmacol. 2014, 156, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Wu, H.; Li, H.; Hu, S.L.; Dai, M.M.; Chen, J. Anti-inflammatory effects and pharmacokinetics study of geniposide on rats with adjuvant arthritis. Int. Immunopharmacol. 2015, 24, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Khan, M.A.; Yang, L.; Yang, M.; Fu, J. Genetic characterization and authentication of Gardenia jasminoides in different regions of China by using improved RAPD analysis. Indian J. Exp. Biol. 2015, 53, 164–169. [Google Scholar] [PubMed]
- Fu, J.; Yang, L.; Khan, M.A.; Mei, Z. Genetic characterization and authentication of Lonicera japonica Thunb. by using improved RAPD analysis. Mol. Biol. Rep. 2013, 40, 5993–5999. [Google Scholar] [CrossRef] [PubMed]
- Desai, P.; Gajera, B.; Mankad, M.; Shah, S.; Patel, A.; Patil, G.; Narayanan, S.; Kumar, N. Comparative assessment of genetic diversity among Indian bamboo genotypes using RAPD and ISSR markers. Mol. Biol. Rep. 2015, 42, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Cheng, J.; Mei, Z.; Zhao, L.; Wei, C.; Fu, S.; Khan, M.A.; Fu, J. Genetic analysis of litchi (Litchi chinensis Sonn.) in southern China by improved random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR). Mol. Biol. Rep. 2015, 42, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Long, Y.; Khan, M.A.; Wei, C.; Fu, S.; Fu, J. Development and significance of RAPD-SCAR markers for Litchi chinensis Sonn. variety authentication by improved RAPD amplification and molecular cloning. Electron. J. Biotechnol. 2015, 18, 35–39. [Google Scholar] [CrossRef]
- Li, S.F.; Tang, S.J.; Cai, W.Q. RAPD-SCAR Markers for Genetically improved NEW GIFT Nile Tilapia (Oreochromis niloticus niloticus L.) and their application in strain identification. Zool. Res. 2010, 31, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Khan, M.A.; Mei, Z.; Yang, M.; Zhang, T.; Wei, C.; Yang, W.; Zhu, L.; Long, Y.; Fu, J. Development of RAPD-SCAR markers for Lonicera japonica Thunb. (Caprifolicaceae) variety authentication by improved RAPD and DNA cloning. Rev. Biol. Trop. 2014, 62, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Xin, G.Z.; Lam, Y.C.; Maiwulanjiang, M.; Chan, G.K.; Zhu, K.Y.; Tang, W.L.; Dong, T.T.; Shi, Z.Q.; Li, P.; Tsim, K.W. Authentication of Bulbus Fritillariae cirrhosae by RAPD-derived DNA markers. Molecules 2014, 19, 3450–3459. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Noormohammadi, Z.; Hasheminejad-Ahangarani, F.Y.; Sheidai, M.; Ghasemzadeh-Baraki, S.; Alishah, O. Genetic diversity analysis in Opal cotton hybrids based on SSR, ISSR, and RAPD markers. Genet. Mol. Res. 2013, 12, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Fu, S.; Khan, M.A.; Zeng, W.; Fu, J. Molecular cloning and development of RAPD-SCAR markers for Dimocarpus longan variety authentication. Springerplus 2013, 2, 501. [Google Scholar] [CrossRef] [PubMed]
- Saju, J.M.; Németh, S.; Szűcs, R.; Sukumaran, R.; Lim, Z.; Wong, L.; Orbán, L.; Bercsényi, M. PCR-based identification of Adriatic specimen of three scorpionfish species (Scorpaenidae, Teleostei). Acta Biol. Hung. 2014, 65, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Khan, M.A.; Mei, Z.; Tania, M.; Yang, L.; Cheng, J. Development of RAPD-SCAR markers for different Ganoderma species authentication by improved RAPD amplification and molecular cloning. Genet. Mol. Res. 2015, 14, 5667–5676. [Google Scholar] [CrossRef] [PubMed]
- Kao, K.W.; Lin, C.Y.; Peng, S.F.; Cheng, Y.M. Characterization of four B-chromosome-specific RAPDs and the development of SCAR markers on the maize B-chromosome. Mol. Genet. Genom. 2015, 90, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Luzio, A.; Coimbra, A.M.; Benito, C.; Fontaínhas-Fernandes, A.A.; Matos, M. Screening and identification of potential sex-associated sequences in Danio rerio. Mol. Reprod. Dev. 2015, 82, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Q.; Wei, G.Y.; Zhou, Y.; Ge, F.; Luo, G.M. Isolation and characterization of twenty-two polymorphic microsatellite markers from Gardenia jasminoides (Rubiaceae). J. Genet. 2014, 93, e22–e24. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.Y.; Wang, X.R.; Zhu, P.L.; Wen, Q.; Yang, C.X. Development of polymorphic microsatellite markers in the medicinal plant Gardenia jasminoides (Rubiaceae). Biochem. Syst. Ecol. 2015, 58, 149–155. [Google Scholar] [CrossRef]
- Fu, J.J. Short Protocols in Medical Molecular Biology; China Medical Science Press: Beijing, China, 2012. [Google Scholar]
- BLAST Algorithm, 2nd ed.An Algorithm for Comparing Primary Biological Sequence Information; National Center of Biotechnology Information: Bethesda, MD, USA, 2002. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 20 March 2015).
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer 3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the G. jasminoides Ellis var. grandiflora Nakai and G. jasminoides are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, Z.; Zhou, B.; Wei, C.; Cheng, J.; Imani, S.; Chen, H.; Fu, J. Genetic Authentication of Gardenia jasminoides Ellis var. grandiflora Nakai by Improved RAPD-Derived DNA Markers. Molecules 2015, 20, 20219-20229. https://doi.org/10.3390/molecules201119687
Mei Z, Zhou B, Wei C, Cheng J, Imani S, Chen H, Fu J. Genetic Authentication of Gardenia jasminoides Ellis var. grandiflora Nakai by Improved RAPD-Derived DNA Markers. Molecules. 2015; 20(11):20219-20229. https://doi.org/10.3390/molecules201119687
Chicago/Turabian StyleMei, Zhiqiang, Boxu Zhou, Chunli Wei, Jingliang Cheng, Saber Imani, Hanchun Chen, and Junjiang Fu. 2015. "Genetic Authentication of Gardenia jasminoides Ellis var. grandiflora Nakai by Improved RAPD-Derived DNA Markers" Molecules 20, no. 11: 20219-20229. https://doi.org/10.3390/molecules201119687