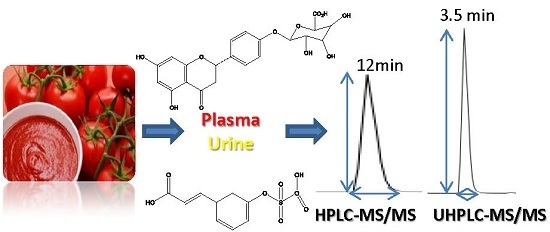
Sensitive and Rapid UHPLC-MS/MS for the Analysis of Tomato Phenolics in Human Biological Samples
Abstract
:1. Introduction
2. Results
2.1. UHPLC-MS/MS Method Development
| Compounds | LOD a (ng/mL) (n = 3) c | LOQ b (ng/mL) (n = 3) c | ||||
|---|---|---|---|---|---|---|
| Formic Acid (0.1%) | Formic Acid (0.05%) | Formic Acid (0.025%) | Formic Acid (0.1%) | Formic Acid (0.05%) | Formic Acid (0.025%) | |
| Caffeic acid | 1.7 ± 0.1 | 2.2 ± 0.1 | 2.1 ± 0.4 | 5.6 ± 0.5 | 7.4 ± 0.1 | 6.8 ± 1.3 |
| 5-Caffeoylquinic acid | 0.8 ± 0.3 | 1.8 ± 0.3 | 5.3 ± 0.4 | 2.7 ± 0.9 | 6.1 ± 0.8 | 17.6 ± 1.3 |
| Dihydrocaffeic acid | 10.7 ± 0.6 | 6.9 ± 0.4 | 6.0 ± 0.2 | 35.7 ± 1.6 | 23.1 ± 0.9 | 19.8 ± 0.7 |
| 3,4-Dihydroxyphenylacetic acid | 2.8 ± 0.3 | 8.2 ± 0.3 | 2.3 ± 0.4 | 9.5 ± 0.8 | 27.3 ± 1.1 | 7.5 ± 1.5 |
| Ferulic acid | 5.5 ± 0.6 | 1.8 ± 0.2 | 1.9 ± 0.4 | 18.3 ± 1.8 | 5.9 ± 0.7 | 6.2 ± 1.1 |
| Hippuric acid | 12.5 ± 1.7 | 9.6 ± 2.5 | 9.4 ± 0.9 | 41.5 ± 5.8 | 32.0 ± 2.1 | 31.5 ± 2.5 |
| Homovanillic acid | n.d. d | n.d. d | 49.2 ± 2.4 | n.d. d | n.d. d | 163.9 ± 6.8 |
| 4-Hydroxyhippuric acid | 0.6 ± 0.1 | 1.3 ± 0.2 | 2.5 ± 0.4 | 2.1 ± 0.6 | 4.2 ± 0.8 | 8.3 ± 1.4 |
| 3-Hydroxyphenylacetic acid | 12.0 ± 1.3 | 7.1 ± 0.3 | 6.6 ± 0.6 | 39.8 ± 3.6 | 23.7 ± 0.7 | 21.8 ± 2.1 |
| 3-(3-Hydroxyphenyl)propionic acid | 5.7 ± 0.9 | 6.6 ± 0.2 | 3.4 ± 0.1 | 18.9 ± 2.7 | 22.0 ± 0.5 | 11.2 ± 0.5 |
| Isoferulic acid | 54.6 ± 2.2 | 5.9 ± 0.1 | 8.9 ± 0.9 | 181.8 ± 8.4 | 19.6 ± 0.4 | 29.7 ± 3.3 |
| Naringenin | 0.5 ± 0.1 | 0.4 ± 0.2 | 0.7 ± 0.4 | 1.6 ± 0.4 | 1.2 ± 0.8 | 2.4 ± 1.1 |
| Phenylacetic acid | n.d. d | n.d. d | 35.3 ± 3.0 | n.d. d | n.d. d | 117.7 ± 8.9 |
| Quercetin | n.d. d | n.d. d | 21.9 ± 0.9 | n.d. d | n.d. d | 73.0 ± 2.3 |
| Quercetin-3-O-glucuronide | n.d. d | n.d. d | 22.4 ± 0.2 | n.d. d | n.d. d | 74.6 ± 0.2 |
| Compounds | Rt (Min) | DP (V) | FP (V) | EP (V) | Quantification Transition | Collision Energy (V) | Confirmation Transition | Collision Energy (V) |
|---|---|---|---|---|---|---|---|---|
| Caffeic acid | 1.18 | −40 | −170 | −10 | 179 → 135 | −20 | 179 → 107 | −30 |
| 5-Caffeoylquinic acid | 1.01 | −50 | −180 | −10 | 353 → 191 | −20 | 353 → 179 | −30 |
| Dihydrocaffeic acid | 1.12 | −50 | −170 | −10 | 181 → 137 | −20 | 181 → 121 | −30 |
| 3.4-Dihydroxyphenylacetic acid | 0.83 | −40 | −170 | −10 | 167 → 123 | −10 | ||
| Ethylgallate (IS) | 1.56 | −60 | −200 | −10 | 197 → 169 | −25 | 197 → 124 | −40 |
| Ferulic acid | 1.70 | −40 | −170 | −10 | 193 → 134 | −20 | 193 → 178 | −30 |
| Hippuric acid | 1.10 | −40 | −170 | −10 | 178 → 134 | −20 | ||
| Homovanillic acid | 1.27 | −40 | −170 | −10 | 181→ 137 | −20 | ||
| 4-Hydroxyhippuric acid | 0.68 | −40 | −170 | −10 | 194 → 100 | −20 | 194 → 150 | −30 |
| 3-Hydroxyphenylacetic acid | 1.28 | −40 | −170 | −10 | 151 → 107 | −10 | ||
| 3-(3-Hydroxyphenyl)propionic acid | 1.67 | −40 | −170 | −10 | 165 → 121 | −20 | 165 → 119 | −35 |
| Isoferulic acid | 1.80 | −50 | −220 | −10 | 193 → 178 | −20 | 193 → 134 | −35 |
| Naringenin | 2.51 | −50 | −190 | −10 | 271 → 151 | −30 | 271 → 119 | −40 |
| Phenylacetic acid | 1.10 | −50 | −170 | −10 | 135 → 91 | −30 | ||
| Quercetin | 2.46 | −60 | −210 | −10 | 301 → 151 | −30 | 301 → 179 | −40 |
| Quercetin-3-O-glucuronide | 1.76 | −60 | −210 | −10 | 477 → 301 | −30 | 477 → 151 | −40 |
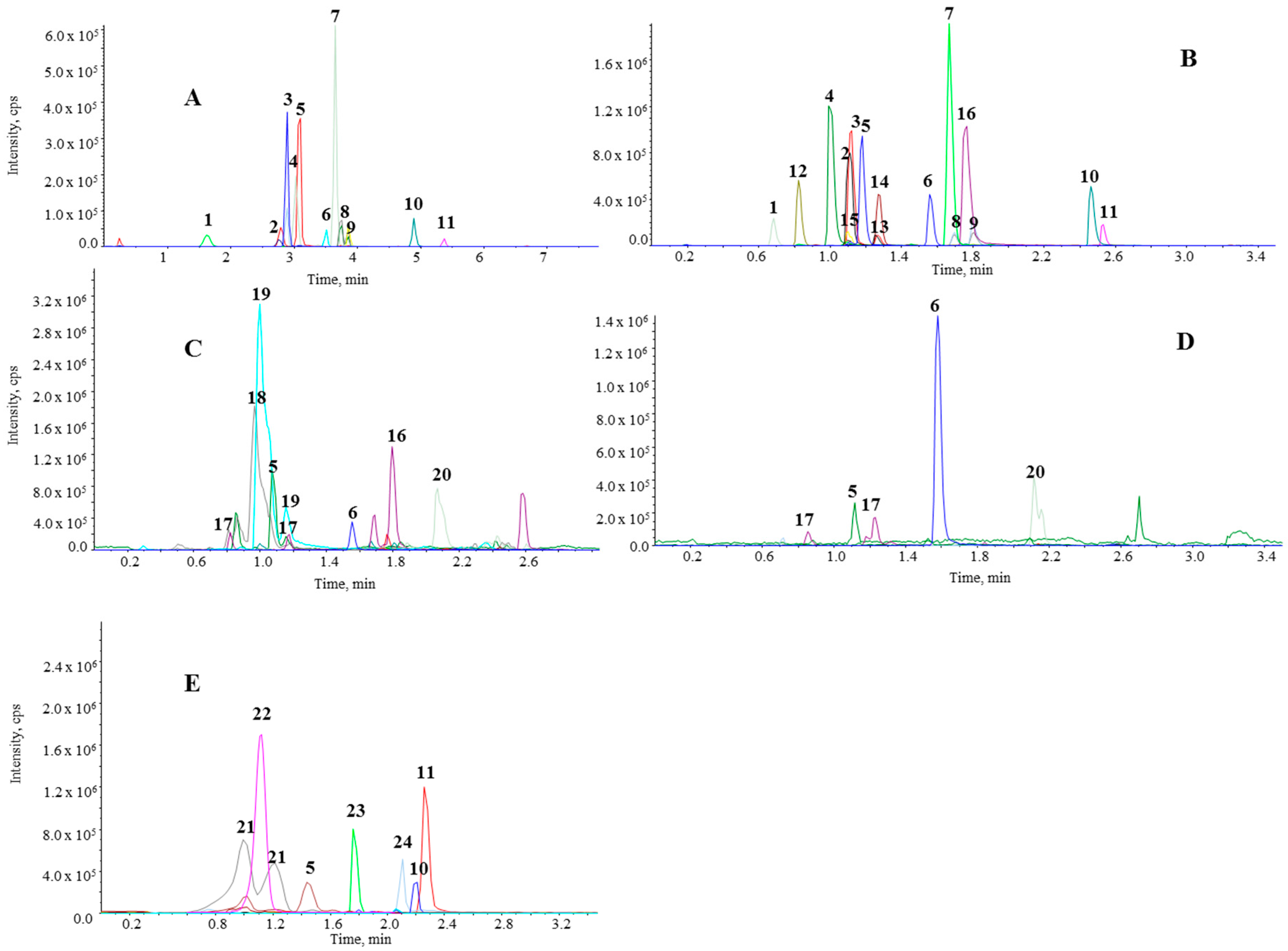
2.2. Validation Parameters
2.2.1. Limits of Detection (LOD) and Quantification (LOQ)
| Compounds | HPLC (n = 3) a | UHPLC (n = 3) a | HPLC (n = 3) a | UHPLC (n = 3) a | HPLC | UHPLC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| URINE | PLASMA | URINE | PLASMA | URINE | PLASMA | URINE | PLASMA | URINE | PLASMA | URINE | PLASMA | |||||
| LOD b (ng/mL) | LOQ c (ng/mL) | LOD b (ng/mL) | LOQ c (ng/mL) | LOD b (ng/mL) | LOQ c (ng/mL) | LOD b (ng/mL) | LOQ c (ng/mL) | Rec. e (%) | Rec. e (%) | Rec. e (%) | Rec. e (%) | Conc. Range f (ng/mL) (r2) g | Conc. Range f (ng/mL) (r2) g | Conc. Range f (ng/mL) (r2) g | Conc. Range f (ng/mL) (r2) g | |
| Caffeic acid | 1.5 ± 0.1 | 6.0 ± 0.1 | 1.8 ± 0.1 | 6.0 ± 0.4 | 0.5 ± 0.2 | 2.0 ± 0.4 | 0.7 ± 0.1 | 2.8 ± 0.1 | 97 ± 4 | 98 ± 3 | 100 ± 2 | 91 ± 2 | 6–3450 (0.990) | 6–3450 (0.993) | 2–1152 (0.994) | 3–1728 (0.995) |
| 5-Caffeoylquinic acid | 0.5 ± 0.1 | 2.0 ± 0.2 | 0.6 ± 0.1 | 2.0 ± 0.1 | 1.1 ± 0.1 | 4.3 ± 0.1 | 0.5 ± 0.1 | 1.7 ± 0.1 | 103 ± 2 | 99 ± 4 | 99 ± 4 | 92 ± 2 | 2–1150 (0.999) | 2–1150 (0.999) | 4–2304 (0.992) | 2–1152 (0.998) |
| Dihydrocaffeic acid | 6.3 ± 0.2 | 25 ± 0.9 | 4.4 ± 0.1 | 15 ± 0.3 | 5.7 ± 0.6 | 20.2 ± 2.1 | 5.3 ± 0.3 | 18.0 ± 1.0 | 101 ± 2 | 104 ± 5 | 97 ± 5 | 90 ± 4 | 25–14,400 (0.993) | 15–14,400 (0.991) | 20–11,520 (0.996) | 18–10,368 (0.992) |
| 3,4-Dihydroxyphenylacetic acid | n.s. d | n.s. d | n.s. d | n.s. d | 1.1 ± 0.1 | 3.5 ± 0.2 | 1.6 ± 0.1 | 5.4±0.2 | n.s. d | n.s. d | 78 ± 3 | 69 ± 2 | n.s. d | n.s. d | 4–2304 (0.994) | 5–2880 (0.998) |
| Ferulic acid | 15 ± 0.7 | 50 ± 2.7 | 18 ± 2.1 | 60 ± 7.0 | 1.9 ± 0.1 | 6.6 ± 0.2 | 1.6 ± 0.1 | 5.6 ± 0.2 | 95 ± 2 | 98 ± 4 | 100 ± 4 | 100 ± 3 | 50–28,800 (0.990) | 60–28,800 (0.996) | 7–4032 (0.998) | 6–3456 (0.990) |
| Hippuric acid | 25 ± 1.6 | 80 ± 3.2 | 25 ± 0.8 | 90 ± 2.1 | 4.4 ± 0.2 | 15.0 ± 0.6 | 2.1 ± 0.3 | 6.5 ± 0.9 | 106 ± 2 | 99 ± 4 | 97 ± 2 | 99 ± 4 | 80–51,840 (0.990) | 90–51,840 (0.992) | 15–8640 (0.999) | 7–4032 (0.991) |
| Homovanillic acid | n.s. d | n.s. d | n.s. d | n.s. d | 4.5 ± 0.1 | 17.3 ± 0.4 | 8.5 ± 0.3 | 29.2 ± 1.0 | n.s. d | n.s. d | 97 ± 4 | 92 ± 4 | n.s. d | n.s. d | 17–9792 (0.996) | 29–16,704 (0.991) |
| 4-Hydroxyhippuric acid | 15 ± 1.2 | 50 ± 2.6 | 15 ± 0.7 | 50 ± 3.2 | 0.6 ± 0.1 | 1.8 ± 0.2 | 1.4 ± 0.1 | 4.6 ± 0.3 | 75 ± 3 | 73 ± 3 | 68 ± 3 | 61 ± 4 | 50–28,800 (0.990) | 50–28,800 (0.992) | 2–1152 (0.999) | 5–2880 (0.999) |
| 3-Hydroxyphenylacetic acid | n.s. d | n.s. d | n.s. d | n.s. d | 0.9 ± 0.1 | 2.8 ± 0.2 | 1.6 ± 0.3 | 5.3 ± 0.9 | n.s. d | n.s. d | 99 ± 2 | 98 ± 2 | n.s. d | n.s. d | 3–1728 (0.999) | 5–2880 (0.998) |
| 3-(3-Hydroxyphenyl)propionic acid | 6.0 ± 0.8 | 20 ± 1.4 | 6.0 ± 1.2 | 20 ± 2.9 | 2.5 ± 0.1 | 9.0 ± 0.2 | 2.4 ± 0.2 | 8.6 ± 0.5 | 97 ± 2 | 99 ± 5 | 98 ± 4 | 94 ± 3 | 20–11,520 (0.995) | 20–11,520 (0.994) | 9–5184 (0.991) | 9–5184 (0.994) |
| Isoferulic acid | 29 ± 4.3 | 90 ± 13.6 | 30 ± 2.4 | 105 ± 8.0 | 2.0 ± 0.3 | 6.2 ± 1.0 | 3.4 ± 0.2 | 12.0 ± 0.5 | 99 ± 2 | 99 ± 4 | 100 ± 3 | 97 ± 4 | 90–28,800 (0.997) | 105–28,800 (0.994) | 6–3456 (0.993) | 12–6912 (0.993) |
| Naringenin | 0.5 ± 0.1 | 2.0 ± 0.1 | 0.5 ± 0.1 | 2.0 ± 0.1 | 0.6 ± 0.1 | 2.8 ± 0.3 | 0.3 ± 0.1 | 1.6 ± 0.1 | 104 ± 4 | 96 ± 3 | 99 ± 2 | 96 ± 2 | 2–1150 (0.999) | 2–1150 (0.995) | 3–1728 (0.999) | 2–1152 (0.999) |
| Phenylacetic acid | n.s. d | n.s. d | n.s. d | n.s. d | 62.5 ± 3.0 | 203.4 ± 9.9 | 44.1 ± 5.4 | 145.8 ± 17.8 | n.s. d | n.s. d | 95 ± 3 | 99 ± 4 | n.s. d | n.s. d | 203–116,929 (0.992) | 146–84,096 (0.994) |
| Quercetin | 1.7 ± 0.1 | 6.0 ± 0.3 | 1.4 ± 0.1 | 5.0 ± 0.4 | 2.3 ± 1.4 | 8.5 ± 4.8 | 4.5 ± 0.1 | 15.8 ± 0.3 | 65 ± 3 | 100 ± 3 | 85 ± 4 | 100 ± 2 | 6–3450 (0.991) | 5–3450 (0.990) | 9–5184 (0.998) | 16–9216 (0.998) |
| Quercetin-3-O-glucuronide | n.s. d | n.s. d | n.s. d | n.s. d | 1.4 ± 0.1 | 4.4 ± 0.1 | 1.9 ± 0.1 | 6.1 ± 0.2 | n.s. d | n.s. d | 99 ± 2 | 95 ± 2 | n.s. d | n.s. d | 5–2880 (0.997) | 6–3456 (0.993) |
2.2.2. Linearity
2.2.3. Recovery
2.2.4. Accuracy and Precision
2.2.5. Stability
2.3. Phenolic Quantification in Tomato Sauce and Biological Samples

| Compounds | Product Ions in MS2 Experiments | Urine (ng/mL) | Plasma (ng/mL) |
|---|---|---|---|
| Caffeic acid * | 179,135,107 | 206 (4.0–822) | n.d. a |
| Caffeic acid glucuronide (CA) | 355,179,135,175,113 | 54.9 (2.0–975) | 6.4 (3.0–12.1) |
| Caffeic acid sulfate (CA) | 259,179,135 | 1396 (10.2–42,286) | 183 (15.3–862) |
| 5-Caffeoylquinic acid * | 353,191,179 | 17.0 (7.2–53.6) | 2.7 (2.0–3.5) |
| Dihydrocaffeic acid * | 181,137,59 | 194.7 (30.1–1031.9) | n.d. a |
| Dihydrocaffeic acid glucuronide (DHCA) | 357,181,137,175,113 | 2127 (23–10,071) | n.d. a |
| Dihydrocaffeic acid sulfate (DHCA) | 261,181,137 | 2775 (52–9424) | n.d. a |
| 3,4-Dihydroxyphenylacetic acid * | 167,123 | 1095 (33.9–2249) | n.d. a |
| Ferulic acid * | 193,134,175 | 453 (31–2139) | 8.1 (6.5–40.3) |
| Ferulic acid glucuronide (FA) | 369,193,134,175,113 | 2852 (110–69,495) | 53.5 (6.7–1881) |
| Ferulic acid sulfate (FA) | 273,193,134 | 22,569 (713–536,479) | 95.5 (24.5–316) |
| Homovanillic acid * | 181,137 | 4050 (1073–9387) | n.d. a |
| 4-Hydroxyhippuric acid * | 194,100 | 33,188 (691–214,695) | n.d. a |
| 3-Hydroxyphenylacetic acid * | 151,107 | 739 (244–1291) | n.d. a |
| 3-Hydroxyphenylacetic acid glucuronide (3-HPAA) | 327,151,107,175,113 | 295 (11.0–1608) | n.d. a |
| 3-Hydroxyphenylacetic acid sulfate (3-HPAA) | 231,151,107 | 16,425 (175–352,967) | n.d. a |
| 3-(3-Hydroxyphenyl)propionic acid * | 165,121 | 1402 (188–4007) | n.d. a |
| 3-(3-Hydroxyphenyl)propionic acid glucuronide (3-(3-HPPA)) | 341,165,121,175,113 | 258 (15.3–4914) | n.d. a |
| 3-(3-Hydroxyphenyl)propionic acid sulfate (3-(3-HPPA)) | 245,165,121 | 3156 (27.6–145,199) | n.d. a |
| Isoferulic acid * | 193,134,175 | 1156 (247–3427) | 108 (10.6–494) |
| Naringenin * | 271,151,119 | 39.1 (6.9–400) | 11.7 (2.0–52.6) |
| Naringenin glucuronide (N) | 854 (214–1558.5) | 73.4 (2.0–830) | |
| Phenylacetic acid * | 135,91 | 1129 (297–6667) | n.d. a |
| Phenylacetic acid glucuronide (PAA) | 311,135,91,175,113 | 318 (206–431) | n.d. a |
| Phenylacetic acid sulfate (PAA) | 215,135,91 | 1378 (110–26,708) | n.d. a |
| Quercetin * | 301,151,179 | 70.8 (9.1–417) | 99.0 (23.7–331) |
| Quercetin glucuronide * | 477,301,151,175,113 | 14.4 (11.9–101) | 20.9 (14.3–63.4) |
| Quercetin sulfate (Q) | 381,301,151 | 32.0 (11.9–489.8) | 3.8 (3.5–4.2) |
3. Discussion
3.1. UHPLC-MS/MS Method Development
3.2. Validation Parameters
3.3. Phenolic Quantification in Tomato Sauce and Biological Samples
4. Experimental Section
4.1. Chemicals
4.2. Method Development
4.2.1. UHPLC Column
4.2.2. Mobile Phase
4.2.3. MS Conditions
4.2.4. Flow Rate and Volume of Injection
4.2.5. Chromatographic Separation
4.3. Quality Parameters
4.4. Method Application: Pilot Dietary Intervention Study
4.4.1. Biological Material
4.4.2. Phenolics of Tomato Sauce and Biological Samples Extraction
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Viuda-Martos, M.; Sanchez-Zapata, E.; Sayas-Barberá, E.; Sendra, E.; Fernández-López, J.; Pérez-Álvarez, J.A. Tomato and tomato byproducts. Human health benefits of lycopene and its application to meat products: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 1032–1049. [Google Scholar] [CrossRef] [PubMed]
- Tresserra-Rimbau, A.; Rimm, E.B.; Medina-Remón, A.; Martínez-González, M.A.; López-Sabater, M.C.; Covas, M.I.; Corella, D.; Salas-Salvadó, J.; Gómez-Gracia, E.; Lapetra, J.; et al. Polyphenol intake and mortality risk: A re-analysis of the PREDIMED trial. BMC Med. 2014, 12, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tresserra-Rimbau, A.; Rimm, E.B.; Medina-Remón, A.; Martínez-González, M.A.; de la Torre, R.; Corella, D.; Salas-Salvadó, J.; Gómez-Gracia, E.; Lapetra, J.; Arós, F.; et al. Inverse association between habitual polyphenol intake and incidence of cardiovascular events in the PREDIMED study. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 639–647. [Google Scholar] [PubMed]
- Pedret, A.; Valls, R.M.; Fernández-Castillejo, S.; Catalán, Ú.; Romeu, M.; Giralt, M.; Lamuela-Raventós, R.M.; Medina-Remón, A.; Arija, V.; Aranda, N.; et al. Polyphenol-rich foods exhibit DNA antioxidative properties and protect the glutathione system in healthy subjects. Mol. Nutr. Food Res. 2012, 56, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Valls-Pedret, C.; Lamuela-Raventós, R.M.; Medina-Remón, A.; Quintana, M.; Corella, D.; Pintó, X.; Martínez-González, M.Á.; Estruch, R.; Ros, E. Polyphenol-rich foods in the Mediterranean diet are associated with better cognitive function in elderly subjects at high cardiovascular risk. J. Alzheimers Dis. 2012, 29, 773–782. [Google Scholar] [PubMed]
- Vallverdú-Queralt, A.; Arranz, S.; Medina-Remón, A.; Casals-Ribes, I.; Lamuela-Raventos, R.M. Changes in phenolic content of tomato products during storage. J. Agric. Food Chem. 2011, 59, 9358–9365. [Google Scholar] [CrossRef] [PubMed]
- Di Lecce, G.; Martínez-Huélamo, M.; Tulipani, S.; Vallverdú-Queralt, A.; Lamuela-Raventós, R.M. Setup of a UHPLC-QqQ-MS Method for the Analysis of Phenolic Compounds in Cherry Tomatoes, Tomato Sauce, and Tomato Juice. J. Agric. Food Chem. 2013, 61, 8373–8380. [Google Scholar] [CrossRef] [PubMed]
- Tulipani, S.; Martinez Huelamo, M.; Rotches Ribalta, M.; Estruch, R.; Ferrer, E.E.; Andres-Lacueva, C.; Illan, M.; Lamuela-Raventós, R.M. Oil matrix effects on plasma exposure and urinary excretion of phenolic compounds from tomato sauces: Evidence from a human pilot study. Food Chem. 2012, 130, 581–590. [Google Scholar] [CrossRef]
- Medina-Remón, A.; Barrionuevo-González, A.; Zamora-Ros, R.; Andres-Lacueva, C.; Estruch, R.; Martínez-González, M.-A.; Diez-Espino, J.; Lamuela-Raventos, R.M. Rapid Folin-Ciocalteu method using microtiter 96-well plate cartridges for solid phase extraction to assess urinary total phenolic compounds, as a biomarker of total polyphenols intake. Anal. Chim. Acta 2009, 634, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Martínez, J.M.; Sanmartin, M.; Rosés, M.; Bosch, E.; Ràfols, C. Determination of dissociation constants of flavonoids by capillary electrophoresis. Electrophoresis 2005, 26, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhao, J.; Chaitep, S.; Guo, Z. Simultaneous analysis of main catechins contents in green tea (Camellia sinensis (L.)) by Fourier transform near infrared reflectance (FT-NIR) spectroscopy. Food Chem. 2009, 113, 1272–1277. [Google Scholar] [CrossRef]
- Epriliati, I.; Kerven, G.; D’Arcy, B.; Gidley, M.J. Chromatographic analysis of diverse fruit components using HPLC and UPLC. Anal. Methods 2010, 2, 1606. [Google Scholar] [CrossRef]
- Vallverdú-Queralt, A.; Rinaldi de Alvarenga, J.F.; Estruch, R.; Lamuela-Raventós, R.M. Bioactive compounds present in the Mediterranean sofrito. Food Chem. 2013, 141, 3365–3372. [Google Scholar] [CrossRef] [PubMed]
- Gruz, J.; Novák, O.; Strnad, M. Rapid analysis of phenolic acids in beverages by UPLC-MS/MS. Food Chem. 2008, 111, 789–794. [Google Scholar] [CrossRef]
- Cao, X.; Wang, C.; Pei, H.; Sun, B. Separation and identification of polyphenols in apple pomace by high-speed counter-current chromatography and high-performance liquid chromatography coupled with mass spectrometry. J. Chromatogr. A 2009, 1216, 4268–4274. [Google Scholar] [CrossRef] [PubMed]
- Magiera, S.; Baranowska, I.; Kusa, J. Development and validation of UHPLC-ESI-MS/MS method for the determination of selected cardiovascular drugs, polyphenols and their metabolites in human urine. Talanta 2012, 89, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Rios, L.Y.; Gonthier, M.-P.; Rémésy, C.; Mila, I.; Lapierre, C.; Lazarus, S.A.; Williamson, G.; Scalbert, A. Chocolate intake increases urinary excretion of polyphenol-derived phenolic acids in healthy human subjects. Am. J. Clin. Nutr. 2003, 77, 912–918. [Google Scholar] [PubMed]
- Ottaviani, J.I.; Momma, T.Y.; Kuhnle, G.K.; Keen, C.L.; Schroeter, H. Structurally related (−)-epicatechin metabolites in humans: Assessment using de novo chemically synthesized authentic standards. Free Radic. Biol. Med. 2012, 52, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Nováková, L.; Solichová, D.; Solich, P. Advantages of ultra performance liquid chromatography over high-performance liquid chromatography: Comparison of different analytical approaches during analysis of diclofenac gel. J. Sep. Sci. 2006, 29, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Ortega, N.; Romero, M.-P.; Macià, A.; Reguant, J.; Anglès, N.; Morelló, J.-R.; Motilva, M.-J. Comparative study of UPLC-MS/MS and HPLC-MS/MS to determine procyanidins and alkaloids in cocoa samples. J. Food Compos. Anal. 2010, 23, 298–305. [Google Scholar] [CrossRef]
- Martínez-Huélamo, M.; Tulipani, S.; Torrado, X.; Estruch, R.; Lamuela-Raventós, R.M. Validation of a new LC-MS/MS method for the detection and quantification of phenolic metabolites from tomato sauce in biological samples. J. Agric. Food Chem. 2012, 60, 4542–4549. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Gao, W.; Phelps, M.A.; Wu, D.; Miller, D.D.; Dalton, J.T. Favorable effects of weak acids on negative-ion electrospray ionization mass spectrometry. Anal. Chem. 2004, 76, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wang, C.; Wang, X.; Xiao, H.; Ma, Q.; Zhang, Q. Comparison of UPLC and HPLC for Analysis of 12 Phthalates. Chromatographia 2008, 68, 803–806. [Google Scholar] [CrossRef]
- Lynch, J. Appendix E: Laboratory Quality Assurance; Official Methods of Analysis of AOAC International: Washington, DC, USA, 2005; Available online: http://www.eoma.aoac.org/app_e.pdf (accessed on 1 July 2015).
- Rubió, L.; Serra, A.; Macià, A.; Borràs, X.; Romero, M.-P.; Motilva, M.-J. Validation of determination of plasma metabolites derived from thyme bioactive compounds by improved liquid chromatography coupled to tandem mass spectrometry. J. Chromatogr. B 2012, 905, 75–84. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.; Pinto, C.B.; Sampaio, G.R.; Yonekura, L.; Catharino, R.R.; Bastos, D.H.M. Development and Validation of Methods for the Extraction of Phenolic Acids from Plasma, Urine, and Liver and Analysis by UPLC-MS. J. Agric. Food Chem. 2013, 61, 6113–6121. [Google Scholar] [CrossRef] [PubMed]
- Urpi-Sarda, M.; Monagas, M.; Khan, N.; Lamuela-Raventos, R.M.; Santos-Buelga, C.; Sacanella, E.; Castell, M.; Permanyer, J.; Andres-Lacueva, C. Epicatechin, procyanidins, and phenolic microbial metabolites after cocoa intake in humans and rats. Anal. Bioanal. Chem. 2009, 394, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Ramešová, Š.; Sokolová, R.; Degano, I.; Bulíčková, J.; Žabka, J.; Gál, M. On the stability of the bioactive flavonoids quercetin and luteolin under oxygen-free conditions. Anal. Bioanal. Chem. 2012, 402, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Slimestad, R.; Fossen, T.; Verheul, M.J. The flavonoids of tomatoes. J. Agric. Food Chem. 2008, 56, 2436–2441. [Google Scholar] [CrossRef] [PubMed]
- Vallverdú-Queralt, A.; Arranz, S.; Casals-Ribes, I.; Lamuela-Raventós, R.M. Stability of the Phenolic and Carotenoid Profile of Gazpachos during Storage. J. Agric. Food Chem. 2012, 60, 1981–1988. [Google Scholar] [CrossRef] [PubMed]
- Helmja, K.; Vaher, M.; Püssa, T.; Raudsepp, P.; Kaljurand, M. Evaluation of antioxidative capability of the tomato (Solanum lycopersicum) skin constituents by capillary electrophoresis and high-performance liquid chromatography. Electrophoresis 2008, 29, 3980–3988. [Google Scholar] [CrossRef] [PubMed]
- Minoggio, M.; Bramati, L.; Simonetti, P.; Gardana, C.; Iemoli, L.; Santangelo, E.; Mauri, P.L.; Spigno, P.; Soressi, G.P.; Pietta, P.G. Polyphenol pattern and antioxidant activity of different tomato lines and cultivars. Ann. Nutr. Metab. 2003, 47, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Rechner, A. Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radic. Biol. Med. 2004, 36, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Caro, G.; Borges, G.; Ky, I.; Ribas, A.; Calani, L.; del Rio, D.; Clifford, M.N.; Roberts, S.A.; Crozier, A. In vitro colonic catabolism of orange juice (poly)phenols. Mol. Nutr. Food Res. 2015, 59, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Stalmach, A.; Williamson, G.; Crozier, A. Impact of dose on the bioavailability of coffee chlorogenic acids in humans. Food Funct. 2014, 5, 1727–1737. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Huélamo, M.; Tulipani, S.; Estruch, R.; Escribano, E.; Illán, M.; Corella, D.; Lamuela-Raventós, R.M. The tomato sauce making process affects the bioaccessibility and bioavailability of tomato phenolics: A pharmacokinetic study. Food Chem. 2015, 173, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.K.; Sharma, N.; Sinha, A.K.; Kumar, N.; Gupta, A.P. Ultrafast UPLC-ESI-MS and HPLC with monolithic column for determination of principal flavor compounds in vanilla pods. J. Sep. Sci. 2009, 32, 3425–3431. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Kord, A. Theoretical and experimental comparison of mobile phase consumption between ultra-high-performance liquid chromatography and high performance liquid chromatography. J. Chromatogr. A 2009, 1216, 6204–6209. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Huélamo, M.; Tulipani, S.; Jáuregui, O.; Valderas-Martinez, P.; Vallverdú-Queralt, A.; Estruch, R.; Torrado, X.; Lamuela-Raventós, R.M. Sensitive and Rapid UHPLC-MS/MS for the Analysis of Tomato Phenolics in Human Biological Samples. Molecules 2015, 20, 20409-20425. https://doi.org/10.3390/molecules201119702
Martínez-Huélamo M, Tulipani S, Jáuregui O, Valderas-Martinez P, Vallverdú-Queralt A, Estruch R, Torrado X, Lamuela-Raventós RM. Sensitive and Rapid UHPLC-MS/MS for the Analysis of Tomato Phenolics in Human Biological Samples. Molecules. 2015; 20(11):20409-20425. https://doi.org/10.3390/molecules201119702
Chicago/Turabian StyleMartínez-Huélamo, Miriam, Sara Tulipani, Olga Jáuregui, Palmira Valderas-Martinez, Anna Vallverdú-Queralt, Ramón Estruch, Xavier Torrado, and Rosa M. Lamuela-Raventós. 2015. "Sensitive and Rapid UHPLC-MS/MS for the Analysis of Tomato Phenolics in Human Biological Samples" Molecules 20, no. 11: 20409-20425. https://doi.org/10.3390/molecules201119702