Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum) from a Genome Survey
Abstract
:1. Introduction
2. Results and Discussion
2.1. Frequency Distribution of Different SSR Markers Types
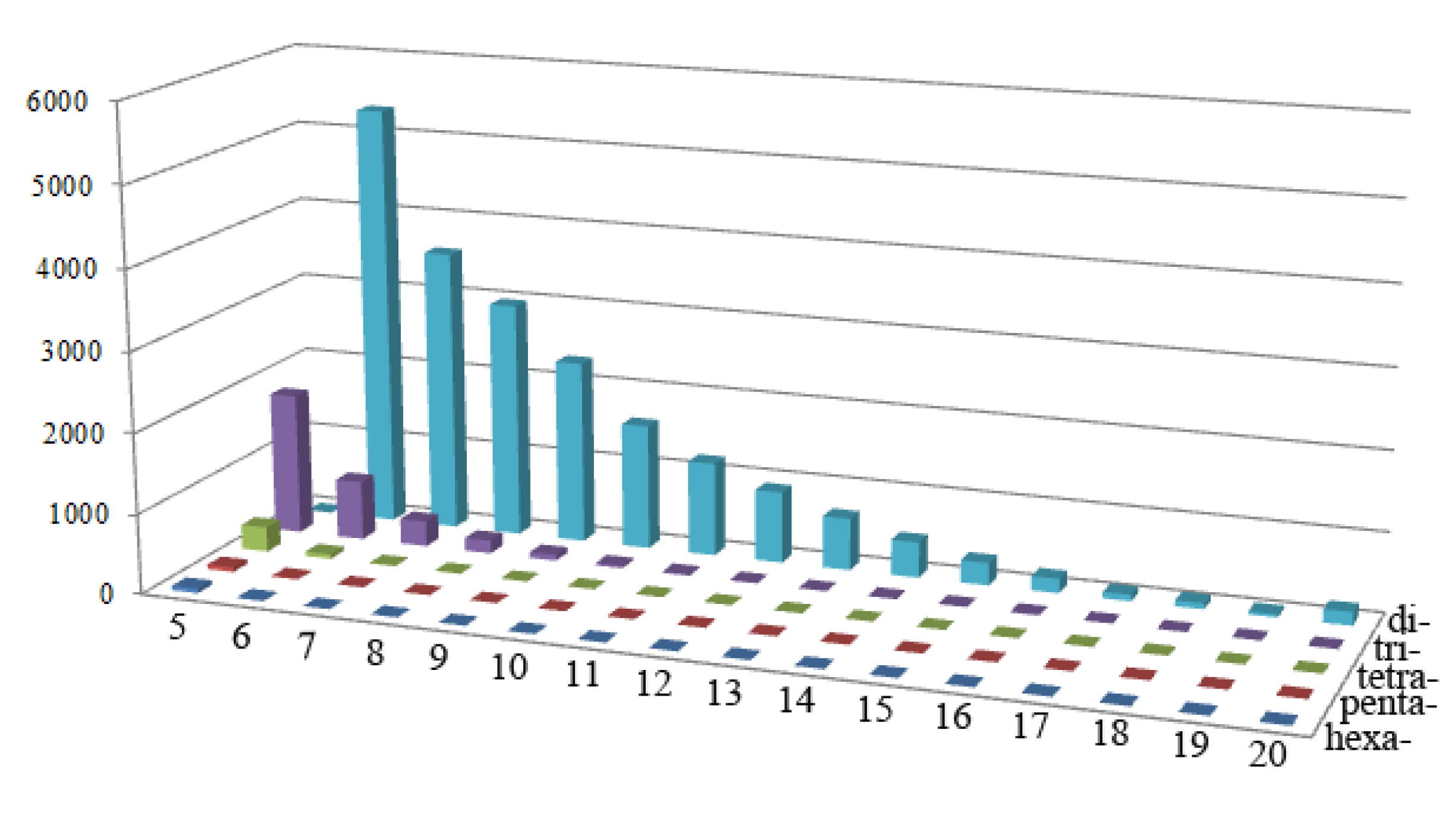
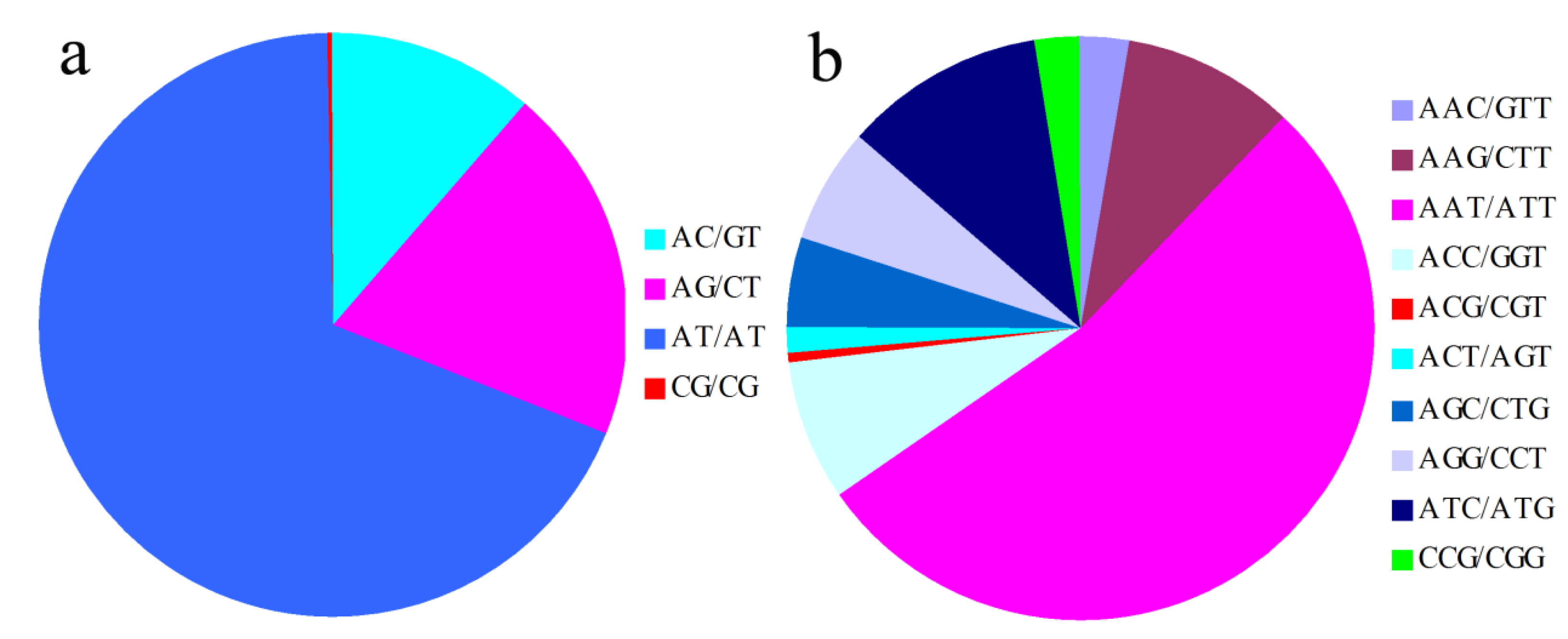
2.2. SSR Marker Polymorphism
2.3. Genetic Relationship Analysis
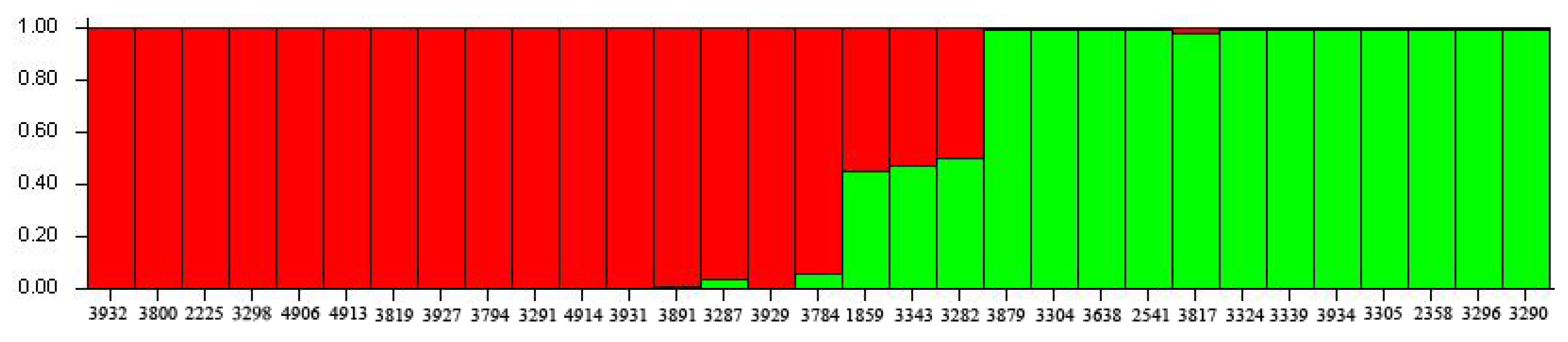
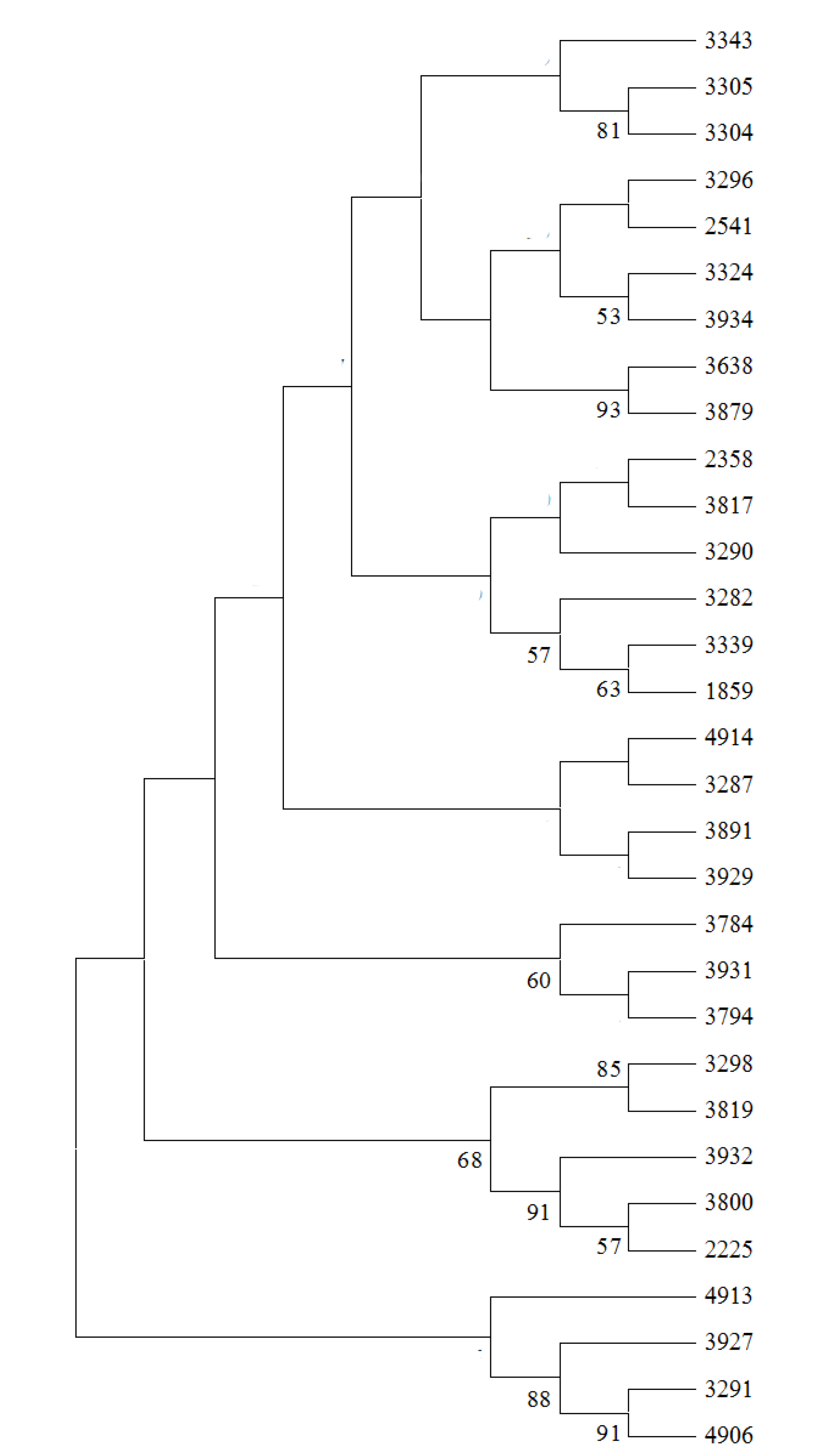
2.4. Core SSR Marker Development

3. Experimental
3.1. Plant Materials, Genome Sequencing, and Genome Survey
3.2. SSR Identification and Primer Design
3.3. PCR and Gel Electrophoresis
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nayar, N.M.; Mehra, K.L. Sesame: Its uses, botany, cytogenetics, and origin. Econ. Bot. 1970, 24, 20–31. [Google Scholar] [CrossRef]
- Johnson, L.A.; Suleiman, T.M.; Lusas, E.W. Sesame protein: A review and prospectus. J. Am. Oil Chem. Soc. 1979, 56, 463–468. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, Y.; Lv, H.; Li, D.; Wang, L.; Zhang, X. Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J. Integr. Plant Biol. 2013, 55, 745–758. [Google Scholar] [CrossRef]
- Yen, G.C. Influence of seed roasting process on the changes in composition and quality of sesame (Sesame indicum) oil. J. Sci. Food Agric. 1990, 50, 563–570. [Google Scholar] [CrossRef]
- Cheung, S.C.; Szeto, Y.T.; Benzie, I.F. Antioxidant protection of edible oils. Plant Foods Hum. Nutr. 2007, 62, 39–42. [Google Scholar] [CrossRef]
- Hirata, F.; Fujita, K.; Ishikura, Y.; Hosoda, K.; Ishikawa, T.; Nakamura, H. Hypocholesterolemic effect of sesame lignan in humans. Atherosclerosis 1996, 122, 135–136. [Google Scholar] [CrossRef]
- Tsai, C.M.; Chen, P.R.; Chien, K.L.; Su, T.C.; Chang, C.J.; Liu, T.-L.; Cheng, H. Dietary sesame reduces serum cholesterol and enhances antioxidant capacity in hypercholesterolemia. Nutr. Res. 2005, 25, 559–567. [Google Scholar] [CrossRef]
- Bhat, K.V.; Babrekar, P.P.; Lakhanpaul, S. Study of genetic diversity in Indian and exotic sesame (Sesamum indicum L.) germplasm using random amplified polymorphic DNA (RAPD) markers. Euphytica 1999, 110, 21–33. [Google Scholar] [CrossRef]
- Ercan, A.G.; Taskin, M.; Turgut, K. Analysis of genetic diversity in Turkish sesame (Sesamum indicum L.) populations using RAPD markers. Genet. Res. Crop Evol. 2004, 51, 599–607. [Google Scholar] [CrossRef]
- Kim, D.H.; Zur, G.; Danin-Poleg, Y.; Lee, S.W.; Shim, K.B.; Kang, C.W.; Kashi, Y. Genetic relationships of sesame germplasm collection as revealed by inter-simple sequence repeats. Plant Breed. 2002, 121, 259–262. [Google Scholar] [CrossRef]
- Hernan, E.L.; Petr, K. Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP). BMC Genet. 2006, 7, 10. [Google Scholar]
- Zhang, Y.X.; Zhang, X.R.; Hua, W.; Wang, L.H.; Che, Z. Analysis of genetic diversity among indigenous landraces from sesame (Sesamum indicum L.) core collection in China as revealed by SRAP and SSR maikers. Genes Genomics 2010, 32, 207–215. [Google Scholar] [CrossRef]
- Wei, L.B.; Zhang, H.Y.; Zheng, Y.Z.; Guo, W.Z.; Zhang, T.Z. Developing EST-derived microsatellites in sesame (Sesamum indicum L.). Acta Agron. Sin. 2008, 34, 2077–2084. [Google Scholar] [CrossRef]
- Watanabe, K.N.; Hirano, R.; Ishii, H.; Oo, T.H.; Gilani, S.A.; Kikuchi, A. Propagation management methods have altered the genetic variability of two traditional mango varieties in Myanmar, as revealed by SSR. Plant Genet. Resour. C 2011, 9, 404–410. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, P.; Choi, Y.; Huang, S.; Gmitter, F. Mining and characterizing microsatellites from citrus ESTs. Theor. Appl. Genet. 2006, 112, 1248–1257. [Google Scholar] [CrossRef]
- Fraser, L.G.; Harvey, C.F.; Crowhurst, R.N.; de Silva, H.N. EST-derived microsatellites from Actinidia species and their potential for mapping. Theor. Appl. Genet. 2004, 108, 1010–1016. [Google Scholar] [CrossRef]
- Jiao, Y.; Jia, H.; Li, X.; Chai, M.; Jia, H. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics 2012, 13, 201. [Google Scholar] [CrossRef]
- Kresovich, S.; Szewc-McFadden, A.K.; Bliek, S.M.; McFerson, J.R. Abundance and characterization of simple-sequence repeats (SSRs) isolated from a size-fractionated genomic library of Brassica napus L. (rapeseed). Theor. Appl. Genet. 1995, 91, 206–211. [Google Scholar]
- Hopkins, M.S.; Casa, A.M.; Wang, T.; Mitchell, S.E.; Dean, R.E.; Kochert, G.D.; Kresovich, S. Discovery and characterization of polymorphic simple sequence repeats (SSRs) in peanut. Crop Sci. 1999, 39, 1243–1247. [Google Scholar] [CrossRef]
- Akkaya, M.S.; Bhagwat, A.A.; Cregan, P.B. Length polymorphisms of simple sequences repeat DNA in soybean. Genetics 1992, 132, 1131–1139. [Google Scholar]
- Wei, W.; Qi, X.; Wang, L.; Zhang, Y.; Hua, W.; Li, D.; Lv, H.; Zhang, X. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 2011, 12, 451. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, L.; Miao, H.; Zhang, T.; Wang, C. Development and validation of genic-SSR markers in sesame by RNA-seq. BMC Genomics 2012, 13, 316. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Wang, Y.W.; Samuels, T.D. Development of 1030 genomic SSR markers in switchgrass. Theor. Appl. Genet. 2011, 122, 677–686. [Google Scholar] [CrossRef]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.K.; Sharma, A.; Singh, V.P.; Gupta, D.K.; Gacche, R.N.; Rana, J.C.; Singh, N.K.; Sharma, T.R. Genome-wide distribution and organization of microsatellites in plants: An insight into marker development in Brachypodium. PLoS One 2011, 6, e21298. [Google Scholar] [CrossRef]
- Cheng, X.; Xu, J.; Xia, S.; Gu, J.; Yang, Y.; Fu, J.; Qian, X.; Zhang, S.; Wu, J.; Liu, K. Development and genetic mapping of mircosatellite markers from genome survey sequences in Brassica napus. Theor. Appl. Genet. 2009, 118, 1121–1131. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Qi, X.; Gao, Y.; Zhang, X. Development and characterization of 59 polymorphic cDNA-SSR markers for the edible oil crop Sesamum indicum (Pedaliaceae). Am. J. Bot. 2012, 99, e394–e398. [Google Scholar] [CrossRef]
- McCouch, S.R.; Cho, Y.G.; Ishii, T.; Temnykh, S.; Chen, X.; Lipovich, L.; Park, W.D.; Ayres, N.; Cartinhour, S. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 2000, 100, 713–722. [Google Scholar] [CrossRef]
- Yermanos, D.M. Sesame. In Hybridization of Crop Plants; Fehr, W.R., Hadley, H.H., Eds.; American Society of Agronomy: Crop Science Society of America: Madison, MI, USA, 1980; pp. 25–36. [Google Scholar]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, I.V. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic asociations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef]
- Dixit, A.; Jin, M.H.; Chung, J.W.; Yu, J.W.; Chung, H.K.; Ma, K.-H.; Park, Y.-J.; Cho, E.-G. Development of polymorphic microsatellite markers in sesame (Sesamum indicum L.). Mol. Ecol. Notes 2005, 5, 736–738. [Google Scholar] [CrossRef]
- Hua, X.; Chang, R.; Cao, Y.; Zhang, M.; Feng, Z.; Qiu, L. Selection of core SSR loci by using Chinese autumn soybean. Sci. Agric. Sin. 2006, 36, 360–366. [Google Scholar]
- Wang, F.; Yi, H.; Zhao, J.; Wang, L.; Sun, Y. Construction of a common PCR program for SSR core primer in maize. J. Maize Sci. 2008, 16, 19–21. [Google Scholar]
- Pan, Z.; Sun, J.; Wang, X.; Jia, Y.; Zhou, Z.; Pang, B.; Du, X.M. Screening of SSR core primers with polymorphism on a cotton panel. Biodivers. Sci. 2008, 16, 555–561. [Google Scholar]
- Li, H.; Jun, Y.; LV, Z.; Yi, B.; Wen, J.; Fu, T.-D.; Tu, J.-X.; Ma, C.-Z.; Shen, J.-X. Screening of Brassica napus core SSR primers. Chin. J. Oil Crop Sci. 2010, 32, 329–336. [Google Scholar]
- Wei, X.; Wang, R.S.; Cao, L.R.; Yuan, N.N.; Huang, J.; Qiao, W.-H.; Zhang, W.-X.; Zeng, H.-L.; Yang, Q.W. Origin of Oryza sativa in China inferred by nucleotide polymorphisms of organelle DNA. PLoS One 2012, 7, e49546. [Google Scholar]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar]
- Kortt, A.A.; Caldwell, J.B.; Lilley, G.G.; Higgins, T.J.V. Amino acid and cDNA sequences of a methionine-rich 2S protein from sunflower seed (Helianthus annuus L.). Eur. J. Biochem. 1991, 195, 329–334. [Google Scholar]
- Bassam, B.J.; Caetano-Anolles, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Liu, K.J.; Muse, S.V. PowerMarker, an integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Hampl, V.; Pavlicek, A.; Flegr, J. Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree, application to trichomonad parasites. Int. J. Syst. Evol. Microbiol. 2001, 51, 731–735. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5, Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure, extensions to linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Sample Availability: Samples are available from the authors’ institute.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wei, X.; Wang, L.; Zhang, Y.; Qi, X.; Wang, X.; Ding, X.; Zhang, J.; Zhang, X. Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum) from a Genome Survey. Molecules 2014, 19, 5150-5162. https://doi.org/10.3390/molecules19045150
Wei X, Wang L, Zhang Y, Qi X, Wang X, Ding X, Zhang J, Zhang X. Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum) from a Genome Survey. Molecules. 2014; 19(4):5150-5162. https://doi.org/10.3390/molecules19045150
Chicago/Turabian StyleWei, Xin, Linhai Wang, Yanxin Zhang, Xiaoqiong Qi, Xiaoling Wang, Xia Ding, Jing Zhang, and Xiurong Zhang. 2014. "Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum) from a Genome Survey" Molecules 19, no. 4: 5150-5162. https://doi.org/10.3390/molecules19045150
APA StyleWei, X., Wang, L., Zhang, Y., Qi, X., Wang, X., Ding, X., Zhang, J., & Zhang, X. (2014). Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum) from a Genome Survey. Molecules, 19(4), 5150-5162. https://doi.org/10.3390/molecules19045150




