Use of EST-SSR Markers for Evaluating Genetic Diversity and Fingerprinting Celery (Apium graveolens L.) Cultivars
Abstract
:1. Introduction
2. Results and Discussion
| Code | Variety | Type | Species |
|---|---|---|---|
| C1 | Xuebaiqincai | Local celery | Apium graveolens L. var. dulce |
| C3 | Jinhuangqincai | Local celery | Apium graveolens L. var. dulce |
| C5 | Jincuifuqin | Local celery | Apium graveolens L. var. dulce |
| C6 | Huangxinqin | Local celery | Apium graveolens L. var. dulce |
| C8 | Shanghaichunqin | Local celery | Apium graveolens L. var. dulce |
| C13 | Tieganqing | Local celery | Apium graveolens L. var. dulce |
| C29 | Wuqiangshiganqincai | Local celery | Apium graveolens L. var. dulce |
| C37 | Shixinqin | Local celery | Apium graveolens L. var. dulce |
| C53 | Duolunshiganqincai | Local celery | Apium graveolens L. var. dulce |
| C111 | Lino | Celery | Apium graveolens L. var. dulce |
| C67 | Jiazhouwang | Celery | Apium graveolens L. var. dulce |
| C87 | Ventura | Celery | Apium graveolens L. var. dulce |
| C114 | Huanghou | Celery | Apium graveolens L. var. dulce |
| C62 | Introduced from USA(fertile) | Celery | Apium graveolens L. var. dulce |
| C63 | Introduced from USA(sterile ) | Celery | Apium graveolens L. var. dulce |
| C65 | - | Celery | Apium graveolens L. var. dulce |
| C68 | Jiahuang | Celery | Apium graveolens L. var. dulce |
| C74 | Dore Golden Spartan | Celery | Apium graveolens L. var. dulce |
| C79 | Kangnaier | Celery | Apium graveolens L. var. dulce |
| C83 | Bailixiqin | Celery | Apium graveolens L. var. dulce |
| C89 | TU52-75 | Celery | Apium graveolens L. var. dulce |
| C97 | Kaifengbolicui | Celery | Apium graveolens L. var. dulce |
| C118 | Guihe | Celery | Apium graveolens L. var. dulce |
| C121 | Qianfang | Celery | Apium graveolens L. var. dulce |
| C123 | Ventura (yellow mutant) | Celery | Apium graveolens L. var. dulce |
| C58 | Majiagouqincai | Middle type | Apium graveolens L. var. dulce |
| C99 | Jinnanhuangxinqin | Middle type | Apium graveolens L. var. dulce |
| C159 | Jinnanshiqin | Middle type | Apium graveolens L. var. dulce |
| C163 | - | Celeriac | Apium graveolens L. var. rapaceum |
| C130 | - | Wild | Apium chilanse |
2.1. Marker Development
| Primer Name | Forward Primer | Reverse Primer |
|---|---|---|
| Fn1 | GCGCTTGGTGTATCTCCACT | AGTGCGTCGAAATATCGCTT |
| Fn2 | GCTTCCGCTGTGTATTTTGA | GGGAAGAAACTGCAACTTGG |
| Fn3 | TGAGCTCCACCAACTGACAC | GCATGAGCAGTTCCAAGACA |
| Fn4 | AATTTACCGCTCTTAGCCTCG | ATAGGCAGAATTTGCGACGA |
| Fn5 | TGAAACCCAAGATCACCCAT | TCATATTGACAGGCAACCGA |
| Fn6 | CCAATCTGGGACTGTGTAAGC | TTCCTGGAGGTGAAGGACTG |
| Fn7 | TGGTGTTGCAGTGTGAATCC | ACCGAAGCATCCTTGAACAG |
| Fn8 | TGGTGTTGCAGTGTGAATCC | ACCGAAGCATCCTTGAACAG |
| Fn9 | CATAGGCTAACGCAGCTTCC | AGTACTCCTTCAGCCGACGA |
| Fn10 | CAGGAGGCTGCAATAACACA | GAGTCGCCGGAATATCAAGA |
| Fn11 | CACACAGACGACTGCTGCTT | ACCATGCATGCTCAACTGAT |
| Fn12 | CACACAGACGACTGCTGCTT | ACCATGCATGCTCAACTGAT |
| Fn13 | CGATCAGGGTACTTGGCAAT | TTTCTATATCCGTCTCATTTCTGTT |
| Fn14 | AACCCTAGCTGCCTCTCTCC | CCATGCCACGAATAGCATTA |
| Fn15 | TGTGTTCTCGCATCTCCAAC | CCAATCTCAACATCGCACAG |
| Fn16 | GTTGGTCAATGCTGCTTCCT | TGTGCCAGGGATACCTTCTC |
| Fn17 | TCACTCACTCCCTTGAGCCT | TGAATCAACACCGTCCATTG |
| Fn18 | CAACCTGAACATCGTTGGTG | TCAACTTGATCTCACGGCAG |
| Fn19 | CTCATACGGTCCAGATCGGT | ATGTCCTGGTGAAGGAGGTG |
| Fn20 | GAGATTGCGATAATGGTGGC | CGCATCACATCACTTAACGG |
| Fn21 | CTGCTCTGAAAGGCTCTGCT | ACAGCTGACATCCTTACCGC |
| Fn22 | TTCACTTGTTCAGCGAGACG | CCTAACCCTAGCTCGTCGTG |
| Fn23 | TCCCATCTCCAATTCCAATC | TTCCTTGCAAGACCATAGGC |
| Fn24 | CATGTCACTGTCGAAGCACC | TGACAATTGCCATTCTCCTG |
| Fn25 | GCCTGAGCATCATAAGAGCC | TATTCACCTTCGTATCCGGC |
| Fn26 | TGTTCCATTATGTGTTGCAGTG | GCGAGATGATGTCAGAACGA |
| Fn27 | ACCATGTCCACCACCTTCTC | GCTGGTTATGGTGGTGCTG |
| Fn28 | ATCGCCAACACCTTCTCAAG | AAGGGTGATTCTGATGGTGC |
| Fn29 | CATATCCAGCACCTCCACCT | TCCAATGGCAACTCACAGAG |
| Fn30 | CTCATTCTTCTTCTTGGCCG | TGCTGAAACGCTACCTCCTT |
| Fn31 | ACATGGAATCTTTCACCTTCA | CATGGCCTAGGAGGAGACAA |
| Fn32 | TTCCTGTCCAGCAGTATCCC | GAATTGAGGGGTGAAAAAGC |
| Fn33 | AAATGAGGTGGTGGTGGAAG | CAATGGGTATGGAATCAGGC |
| Fn34 | AGTACGGTGTCTACGACGGG | CCTCCACCATGATTACCACC |
| Fn35 | CATACTTCTTTGGGGGCTCA | ACACAAACTTTCGGCCAGAT |
| Fn36 | AAGGTCAAGGTCCTGTGGTG | GGTTTAGGCCTCCAATAGCC |
| Fn37 | ACAGTACGTGTCTCCCCCTG | AACAACCCTATGATGGCTGC |
| Fn38 | GTTTGAGCCTCCGCTTACAG | TGCCAGTGACACTCTTCACC |
| Fn39 | GTGACGAAGGAATTGACCGT | ATTTGTTGTCGGGTTCCAAA |
| Fn40 | CTGGCACTTGTACGAAACCA | TATGGGCTGTTGATGACAGG |
| Fn41 | TTCAACCCAGACTTCAACCC | GCAGCCTTCAAATCCAGTTC |
| Fn42 | CCCAGCCCTATCAATCTTCA | CCCCTGCCAAGTCTGTTAAT |
| Fn43 | GAGACAGAGACCATGGGGAA | CGGTTTCGGTTTCGATTTTA |
| Fn44 | TCTTGTCCATTAAAAATGTACCCA | TGCGCATAATGAAAGGATCA |
| Fn45 | AGCAGCACAACAACACTTGG | TTAGGGTCTCTTGTCGAGGC |
| Fn46 | GCAAGTTACCACCCCAGAAA | CCTTTTTCAAAAGCTTCCCC |
| Fn47 | AATTGGCCAGAGCAGAGAAA | TCCTTTATCCCTGACCATGC |
| Fn48 | AATGAGGTTGTTTTTCCCCC | GTAAAGGCCCAAACTCCTCC |
| Fn49 | ACAACAATTTCAGGGCCAAG | TCTTGATATCGGCTTCCTGG |
| Fn50 | ACATTTGTTGCTAGGGTGGC | GCACGAATAGCCGTCCTAAA |
| Fn51 | TCCAATCTCCGGAGCAATAC | GCGGTGGACGAGTAAAAGAG |
| Fn52 | AAACCACCAAACAAGGTGCT | TGAAGTGGAGGAGCAGGAGT |
| Fn53 | ATTCCCAGATGGCTGCATAG | AATTCCAGCAAGCTCAAGGA |
| Fn54 | CCCTCTCCCTATCTTCCTCG | TGAGATTGACTCGGTTGCTG |
| Fn55 | AAAGAAGAAACGGGGATGCT | AACAGCAAGCAGTTCAGTAGTCA |
| Fn56 | CTACACCGCCAATTCAACCT | TAAGCATACACCCCCTCACC |
| Fn57 | TAATGGTGGAAAGAAAGGCG | GGCATACCACTCATTTGGCT |
| Fn58 | CCTAGGCGAACTCTCCTCCT | GGGAATGATCCTCCTTCCAT |
| Fn59 | AGAGGGATAAAATCCCGGTG | TGGAAATGCAAAAGAAGCAA |
| Fn60 | CCACCACCACAACTACAACG | AAGCCGAGTAATGCTGGAAA |
| Fn61 | AGGGTTCGTCGGTTGTAGTG | TCCCCATGTCTATTCTTCGC |
| Fn62 | TCTCGACGAGTTTTCACCTG | GGTCTCTTTGGTGCCATTGT |
| Fn63 | TCAATTAGATCCAGACGCCC | TCTTCTGCCTCCTCTTCGAG |
| Fn64 | CTGTTTTCTCCCTCGTCTGC | CCCCATCTCTGCTATAGCTCC |
| Fn65 | CTTTCTGGGTAAGAAGGCCC | CCAACCCACAACGTCTTACC |
| Fn66 | TCATTGGACAAACAGGACGA | CTGTTTGGCGCTCAATTTTT |
| Fn67 | TACATTTGTGGGATTGGGGT | CCGCCAAAACATTGACAGTA |
| Fn68 | TCAGCTAAGCCACCCTGATT | GTTGCTGCTGGAGAAAGGAG |
| Fn69 | TCCATGAATCTTTCAAGCCC | TCCAAAGTCCAATCCCATTT |
| Fn70 | TAACTGAGTCGGTTGGGTCC | CCTCTCTTTTACCAGCCAAGC |
| Fn71 | TCAACACTCAATTTAAACACCCA | TGATAACGATCGTGACGGAA |
| Fn72 | CATCAACACTCGAAATCGAAA | CAAGATGCTTGTTATCCTTGCT |
| Fn73 | GGATCGGAGGAAGGAAGAAA | GGAGGTGGAGGAGGAGAGTT |
| Fn74 | CCCCCAAACAATAAGTATCCC | TTGGAACTTTTTGTGTCCATTG |
| Fn75 | GCCAGCAGTGTCCCATATTT | GCCCGGAAAAATAACAATCA |
| Fn76 | TTCTACCACTTTCCTTGAATCC | CAGGAGCAGTCTCGATTTCC |
| Fn77 | GGTGTATTTGAATATTAACACCTTTCG | AGAGATGGTGGTCTTGTGGG |
| Fn78 | ACAAGCCCCCTCTACTTTGG | ATGTTGCCAGTTCAGGTTCC |
| Fn79 | TGGGACCCATTTCTTGATTT | AAAATTGCTCCGATTTGTGC |
| Fn80 | CCAGGTAAGCCCAGTCTCAA | TTTTTCTCAATTAAAACTTGCTCATTT |
| Fn81 | AATCCTTGAACTAACCGGGG | CTCTTCGCCACCAGATTCAT |
| Fn82 | GGACGCCCAAGAGAGGTAGT | AGTGGTCTCGACATTTTCCC |
| Fn83 | CCACACCTTGATCGTTGAGA | TTGCTTCTTCCGGCTCTTTA |
| Fn84 | GTTACTTGACGGCACCGTTT | ATCAGTTCTTCATCCGTGGG |
| Fn85 | TCACCCTCTCATCCACATCA | GCAGTGGGTGGATCTAGGAA |
| Fn86 | TCAAATGGACGACGAATCAA | TGCAATGATTTATCCCCCTC |
| Fn87 | CACACACAGGACACACATATTTC | GTAAAGCCGTCTTGGACGAG |
| Fn88 | CGGCATCTTCTCTTCCTCAC | TGTTTGGATCTTTTCTGTTTTCA |
| Fn89 | CAGAAGCGGCTCCTTCTCTA | CCCATTTGAGCTTCACCACT |
| Fn90 | CTAAACGACGCCGTTACCAT | GCTTCTCTCCGCCTTGTATG |
| Fn91 | GGCATACATCGGACGCTAAT | TTGACCCTTTATCTCAATACACACA |
| Fn92 | CCCTCTCTCTCCCTCCTGTC | ACGATTAGCCATTGGTGAGC |
| Fn93 | TGTGTGCTGATTTGAAACCC | ACCGACACTCCACCTTCATC |
| Fn94 | CACCTCTGCTTTCACGGAAC | GTCCAAGAGTGGTCCTCACC |
| Fn95 | ATGGTAACACCACCCTGGAA | GCTTCAACCAGGCAAAGACT |
| Fn96 | TACTTACACCCCTCCCTCCC | TGCAGCACAAGGGATTCATA |
| Fn97 | AAGAGCGATCAAGAACAGGG | TCCCATCTCTCTCCCTCGTA |
| Fn98 | TGCGAACAATACAGTCCCAA | CAGATCCAAACACAGAATTAGCA |
| Fn99 | GAAGAAAGAGGAGAAGGCCG | TCTCGAAACCACCCATCTTC |
| Fn100 | GCGATCCCTAATCAATCCAA | CTTTGAGAGTTACGACGGGC |
| Fn101 | TCAATGGTGTAGAACCAGAACAA | CCCAGATGCTTAAAAGAACCA |
| Fn102 | ACAGGAGGCACTGGTCTCAC | CATTAAAATCCCACAAAAACTTCA |
| Fn103 | TCCCATTCCATTTCAACCTC | AGAGGTGTGGGGAGATTGTG |
| Fn104 | GCGGGGACACTCCACTACT | TTGATCATCAGCAGACTGGC |
| Fn105 | CAAAAATTTAACCCCATACCC | ACATGTACGGACGTTGTGGA |
| Fn106 | GCTTTGCACACACACACACA | CTTTCCCTCGACCTCATCCT |
| Fn107 | GATTTTTCCGATGCAGCACT | GGCATGCACCAAACGTTATC |
| Fn108 | GAGGAGGCTGTTACGTGGAG | TCCCTTTTCTCACTCCATTCC |
| Fn109 | GTAGAAGGCCTGCAGATGCT | GTCTTTGCTTCTCCTCACCG |
| Fn110 | GCACCAGCAAGAGGAGACTT | TTGTTGCTTGCCAGTGAAAC |
| Fn111 | AAGCGAGTAGCTGAAAGGCA | CACTACCACCTCCGATTGCT |
| Fn112 | CCAAGCTTCGACCATTGTTT | TTGTACATCGGTGAAACGGA |
| Fn113 | AGCAGAAAGGCGTTCCACTA | GTTGAGCCCTTCCTGCATAA |
| Fn114 | CGCCCTCTTCCTTTATCTCC | CACTTAGGTTTACCGCTGCC |
| Fn115 | CACGTTTGGTGACATTCCAG | ACAATTATCTCCTTCCGCCC |
| Fn116 | TCCTCTCCTCACCAAACCAC | CACAACCCTTCAACATCACG |
| Fn117 | GTGGTTGGTGGGGATCATAG | GCCCAAAGTTCTTCCAAACA |
| Fn118 | CAATCATATTAATCATCCCCAAA | GAGTTGGTCTGCAGGAGGAG |
| Fn119 | TAAGATGCATGAGGCACGAC | GACTTTGATGCGCACTTTCA |
| Fn120 | TGATTTGTGCACCAAAAGGA | GGAGAGTCGACCGATTCAGA |
| Fn121 | AACTCAGCAACCGGAATCAC | ATACGTAGACGCATCGGAGG |
| Fn122 | GCACACAATAAGCCTCCCAT | CACATGCTACAAAACAGGCG |
| Fn123 | CCACTGGACATTTCTTGGCT | TTTACAAGCCCCAACAGAGC |
| Fn124 | CTGGAACCGGAGTAGGTGAA | AACAGCCTTTACCCTTCATCA |
| Fn125 | ATCTGCCTGTAGCCGAACAG | CTCTTAGTTGGCGCTGCTCT |
| Fn126 | ATAATTTGCCCAACGCTCTG | CTCTCTTGAAAAACACGGGC |
| Fn127 | CAACACAAACACCAAAACCCT | CGTGCCTCATTGGGTTCTAT |
| Fn128 | GTTGTACTTGGTGCGGAGGT | CAAAATTCCAAAAGCCCAAA |
| Fn129 | TCTTTCGATTTGGATTTGGG | TAGAGCTCTCGGCCTCTTCA |
| Fn130 | TTGGTGCCATTGTTGTTGTT | AACGCCTTTCTTCCCAATTT |
| Fn131 | CACCGCGATTCTTCTCTCTC | CGACATCGTCTCTCTCCCTC |
| Fn132 | TTCTTTTTCTGTTCCGCCC | CCGCCGTTAGAGACAAACTC |
| Fn133 | ATTGAAACCCCACCACTGAA | AACGGCCAGAAAAAGCTGTA |
| Fn134 | TGGTTGGGGGAGAATTGTAA | TGAGTTTGCCACAACTGACA |
| Fn135 | TCCCGATAACAAGAGAGAGACT | TGGAGATGAACAAGGGAAGG |
| Fn136 | AGTCCTCAGTTCTCCTGGCA | CAGAATGGTGATGCTGATGG |
| Fn137 | CCAGGACATACATACGTTCTCAA | GACGGACTTAGCCCCCTTAT |
| Fn138 | TAGCTGCGGTTGATTCAGTG | ATTATCAGCGGAAGGCACAC |
| Fn139 | TGCACCACCAAAAACACCTA | GAGGAGGGGTTGAGTGATGA |
| Fn140 | TCACCACCCCTAATTACCGA | AGATAAACCGGGGAGCTTGT |
2.2. Marker Informative Analysis of Accessions
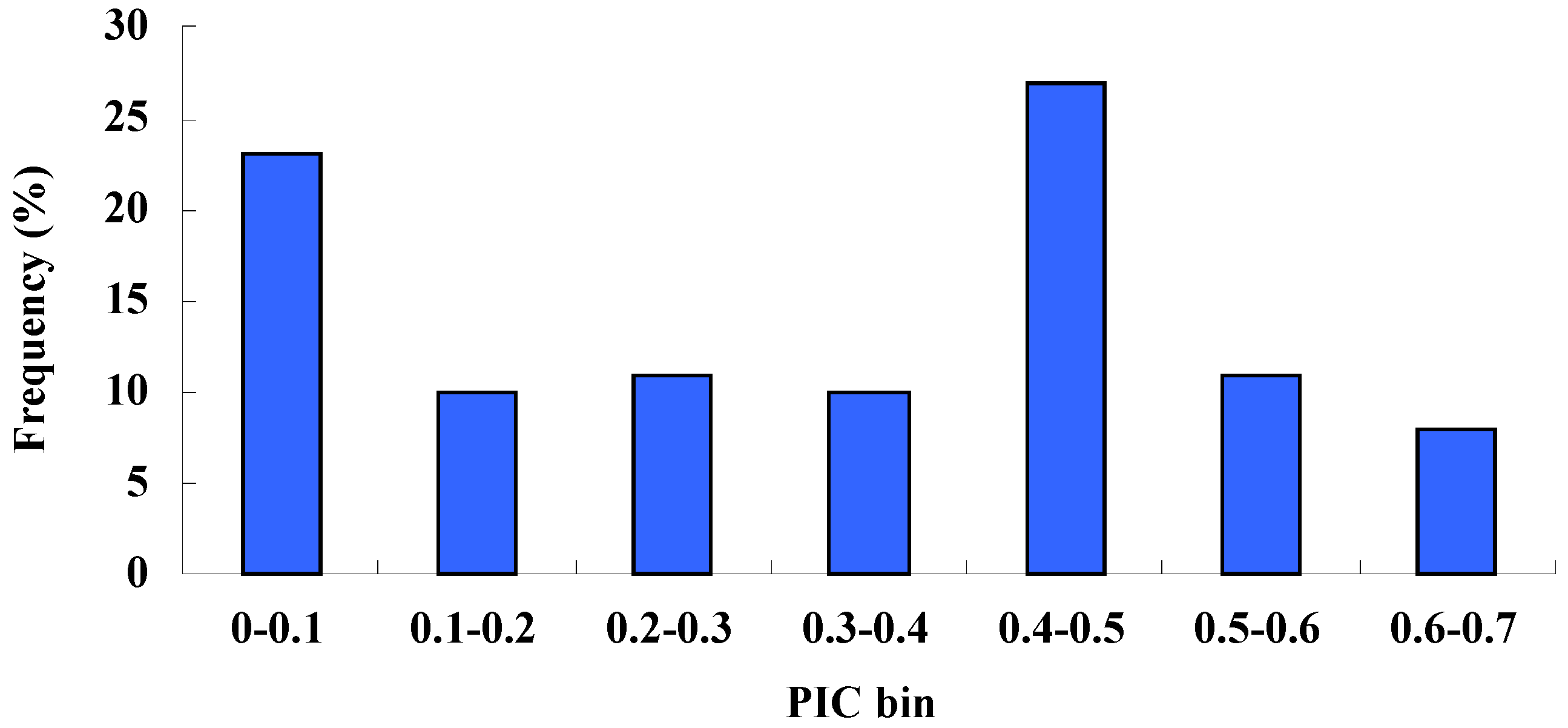
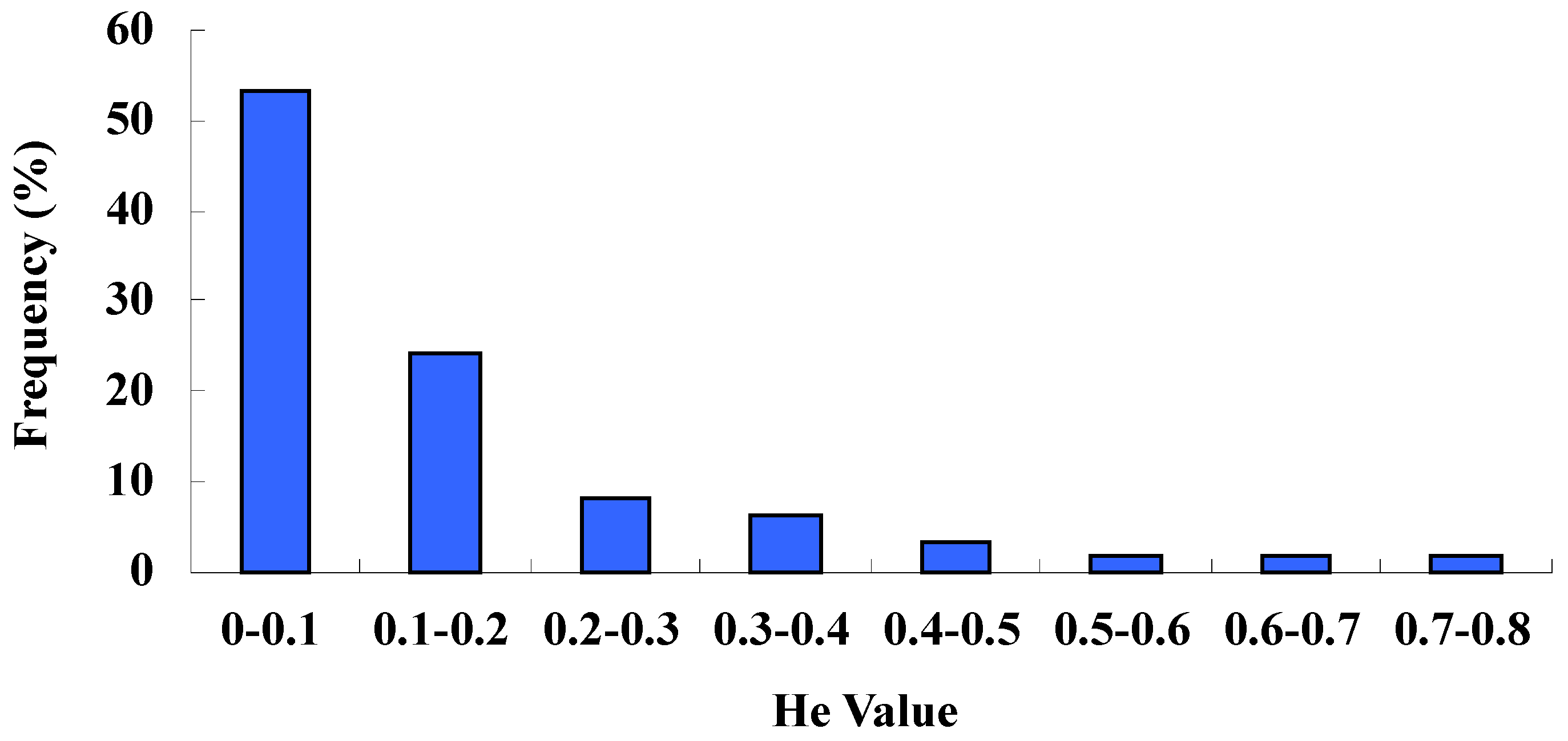
2.3. Analysis of Molecular Variance (AMOVA)
| Source of variation | Percentage of variation | p-value |
|---|---|---|
| Among horticultural types | 22% | 0.001 |
| Among Individuals | 43% | 0.001 |
| Within Individuals | 35% |
| Horticultural type | Local celery | Celery | Middle type |
|---|---|---|---|
| Local celery | - | ||
| Celery | 0.231 | - | |
| Middle type | 0.086 | 0.261 | - |
2.4. PCA Analysis

2.5. Genetic Distance Analysis
2.6. Cluster and Population Structure Analysis
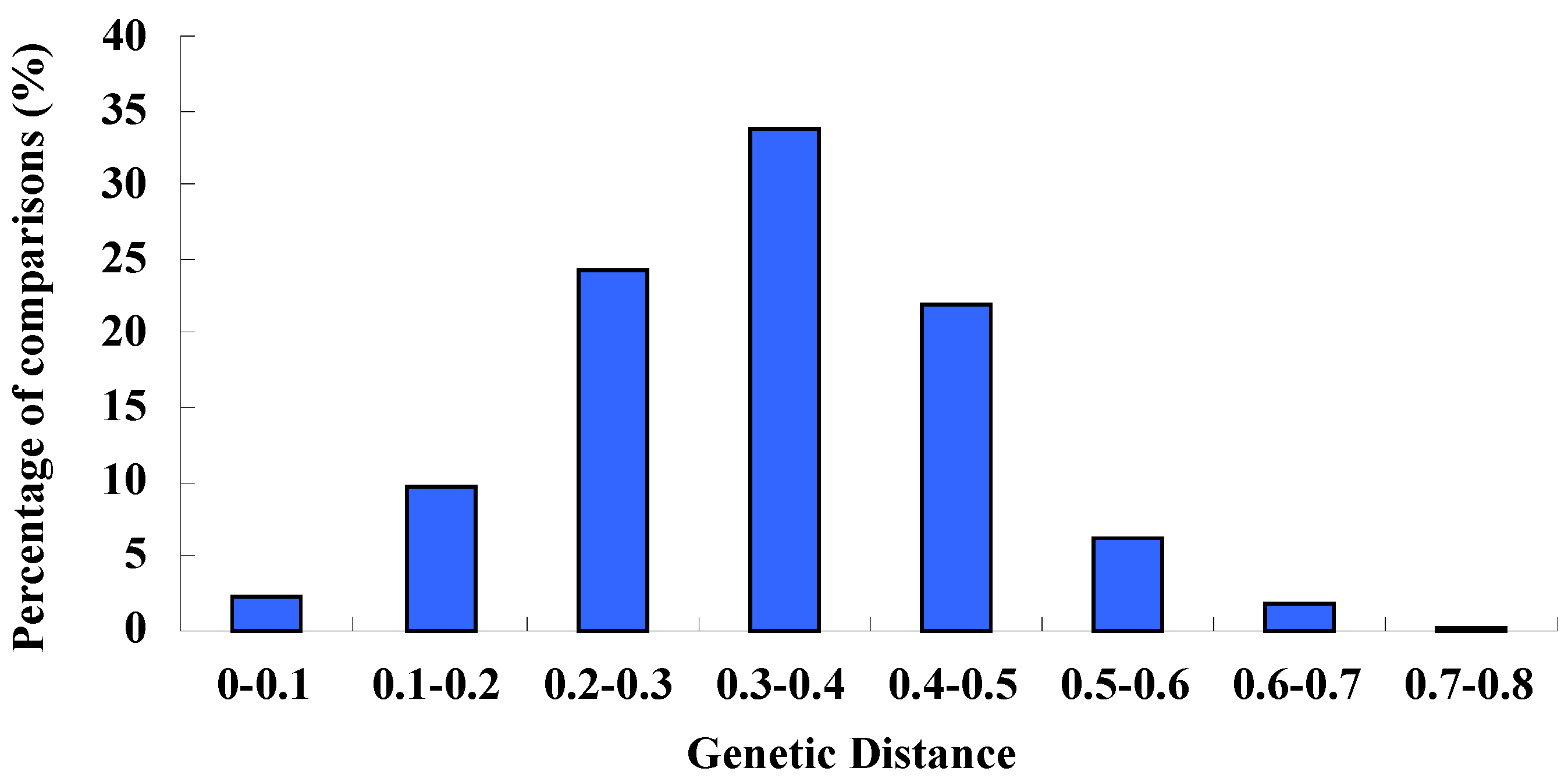

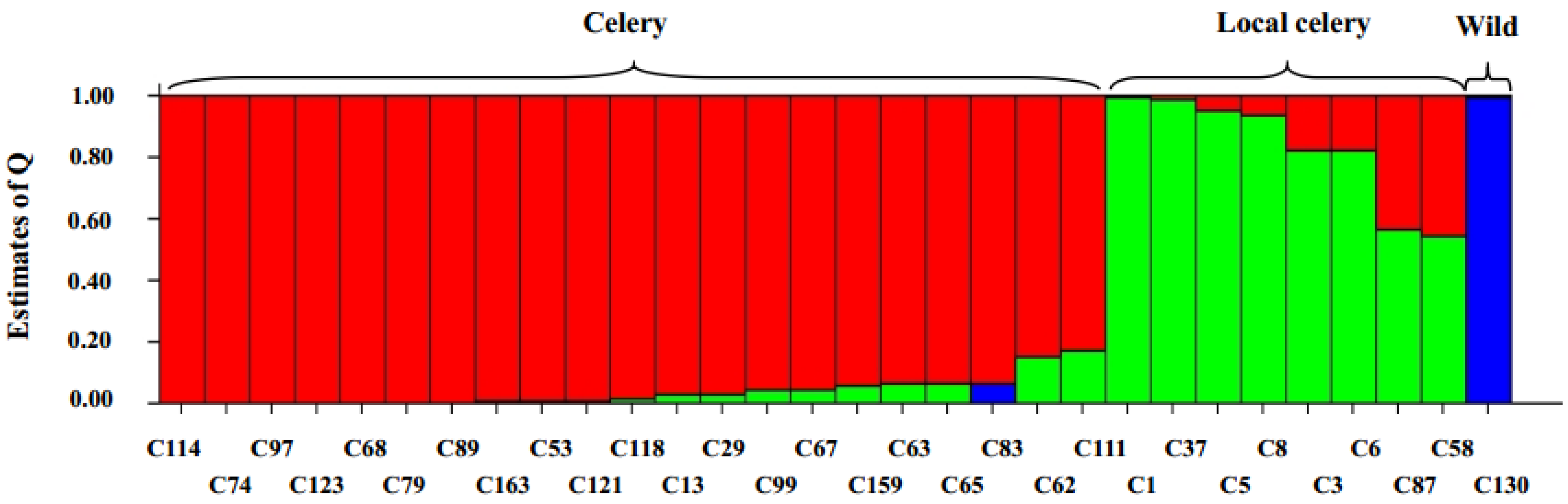
2.7. Genotypic Diversity

3. Experimental
3.1. Plant Materials and DNA Isolation
3.2. Development of Genic SSR Markers and Genotyping with Markers
3.3. Analysis of Marker Polymorphism and Genetic Heterozygosity
3.4. AMOVA and PCA Analysis
3.5. Genetic Diversity and Population Structure Analysis
3.6. Identification of Genotypes
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Arus, P.; Ortan, T. Inheritance patterns and linkage relationships of eight genes of celery. J. Hered. 1984, 75, 11–14. [Google Scholar]
- Huestis, G.; McGrath, J.M.; Quiros, C.F. Development of genetic markers in celery based on restriction fragment length polymorphisms. Theor. Appl. Genet. 1993, 85, 889–896. [Google Scholar]
- Yang, X.; Quiros, C. Identification and classification of celery cultivars with RAPD markers. Theor. Appl. Genet. 1993, 86, 205–212. [Google Scholar]
- Domblides, A.; Domblides, H.; Kharchenko, V. Discrimination between celery cultivars with the use of RAPD markers. Proc. Latv. Acad. Sci. B 2008, 62, 219–222. [Google Scholar]
- Li, G.; Quiros, C.F. Use of amplified fragment length polymorphism markers for celery cultivar identification. HortScience 2000, 35, 726–728. [Google Scholar]
- Ju, J. An analysis of celery genetic diversity by AFLP. Chin. Agric. Sci. Bull. 2007, 23, 120–123. [Google Scholar]
- Wang, S.; Yang, W.; Shen, H. Genetic diversity in Apium graveolens and related species revealed by SRAP and SSR markers. Sci. Hortic. 2011, 129, 1–8. [Google Scholar] [CrossRef]
- Ince, A.; Karaca, M.; Turgut, K. Development of new set of EST-SSR primer pairs for celery (Apium graveolens L.). Planta Med. 2010, 76, P036. [Google Scholar]
- Qing-kuo, L.; Ou-jing, L.; Ji-bo, Z.; Xin, Z.; Zhu, Z.; Rui, C.; Shi-yong, X.; Yong, W.; Yong-ze, G. Construction of SSR-based molecular fingerprinting and analysis of genetic diversity for celery varieties from Tianjin. Tianjin Agric. Sci. 2012, 18, 7–11. [Google Scholar]
- Yu, M.; Chu, J.; Ma, R.; Shen, Z.; Fang, J. A novel strategy for the identification of 73 Prunus domestica cultivars using random amplified polymorphic DNA (RAPD) markers. Afr. J. Agric. Res. 2013, 8, 243–250. [Google Scholar]
- Zhang, J.; Shu, Q.; Liu, Z.; Ren, H.; Wang, L.; de Keyser, E. Two EST-derived marker systems for cultivar identification in tree peony. Plant Cell Rep. 2012, 31, 299–310. [Google Scholar] [CrossRef]
- Figueiredo, E.; Canhoto, J.; Ribeiro, M. Fingerprinting and genetic diversity of Olea europaea L. ssp. Europaea accessions from the cultivar Galega using RAPD markers. Sci. Hortic. 2013, 156, 24–28. [Google Scholar] [CrossRef]
- Hameed, U.; Pan, Y.; Muhammad, K.; Afghan, S.; Iqbal, J. Use of simple sequence repeat markers for DNA fingerprinting and diversity analysis of sugarcane (Saccharum spp) cultivars resistant and susceptible to red rot. Genet. Mol. Res. 2012, 11, 1195–1204. [Google Scholar] [CrossRef]
- Baldwin, S.; Pither-Joyce, M.; Wright, K.; Chen, L.; McCallum, J. Development of robust genomic simple sequence repeat markers for estimation of genetic diversity within and among bulb onion (Allium cepa L.) populations. Mol. Breed. 2012, 30, 1401–1411. [Google Scholar] [CrossRef]
- Park, Y.-J.; Lee, J.K.; Kim, N.-S. Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules 2009, 14, 4546–4569. [Google Scholar] [CrossRef]
- Dos Santos, J.M.; Duarte Filho, L.S.C.; Soriano, M.L.; da Silva, P.P.; Nascimento, V.X.; de Souza Barbosa, G.V.; Todaro, A.R.; Ramalho Neto, C.E.; Almeida, C. Genetic diversity of the main progenitors of sugarcane from the RIDESA germplasm bank using SSR markers. Ind. Crops Prod. 2012, 40, 145–150. [Google Scholar] [CrossRef]
- Zhang, W.W.; Pan, J.S.; He, H.L.; Zhang, C.; Li, Z.; Zhao, J.L.; Yuan, X.J.; Zhu, L.H.; Huang, S.W.; Cai, R. Construction of a high density integrated genetic map for cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2012, 124, 249–259. [Google Scholar] [CrossRef]
- Schouten, H.J.; van de Weg, W.E.; Carling, J.; Khan, S.A.; McKay, S.J.; van Kaauwen, M.P.; Wittenberg, A.H.; Koehorst-van Putten, H.J.; Noordijk, Y.; Gao, Z. Diversity arrays technology (DArT) markers in apple for genetic linkage maps. Mol. Breed. 2012, 29, 645–660. [Google Scholar] [CrossRef]
- Qin, H.; Feng, S.; Chen, C.; Guo, Y.; Knapp, S.; Culbreath, A.; He, G.; Wang, M.L.; Zhang, X.; Holbrook, C.C. An integrated genetic linkage map of cultivated peanut (Arachis hypogaea L.) constructed from two RIL populations. Theor. Appl. Genet. 2012, 124, 653–664. [Google Scholar] [CrossRef]
- Moriguchi, Y.; Ujino-Ihara, T.; Uchiyama, K.; Futamura, N.; Saito, M.; Ueno, S.; Matsumoto, A.; Tani, N.; Taira, H.; Shinohara, K. The construction of a high-density linkage map for identifying SNP markers that are tightly linked to a nuclear-recessive major gene for male sterility in Cryptomeria japonica D. Don. BMC Genomics 2012, 13, 95. [Google Scholar] [CrossRef]
- Sinha, P.; Tomar, S.; Singh, V.K.; Balyan, H. Genetic analysis and molecular mapping of a new fertility restorer gene Rf8 for Triticum timopheevi cytoplasm in wheat (Triticum aestivum L.) using SSR markers. Genetica 2013, 141, 431–441. [Google Scholar]
- Rauscher, G.; Simko, I. Development of genomic SSR markers for fingerprinting lettuce (Lactuca sativa L.) cultivars and mapping genes. BMC Plant Biol. 2013, 13, 11. [Google Scholar] [CrossRef]
- Raman, H.; Raman, R.; Eckermann, P.; Coombes, N.; Manoli, S.; Zou, X.; Edwards, D.; Meng, J.; Prangnell, R.; Stiller, J. Genetic and physical mapping of flowering time loci in canola (Brassica napus L.). Theor. Appl. Genet. 2013, 126, 119–132. [Google Scholar] [CrossRef]
- Chung, J.-W.; Kim, T.-S.; Suresh, S.; Lee, S.-Y.; Cho, G.-T. Development of 65 Novel Polymorphic cDNA-SSR markers in Common Vetch (Vicia sativa subsp. sativa) using next generation sequencing. Molecules 2013, 18, 8376–8392. [Google Scholar] [CrossRef]
- Mudalkar, S.; Golla, R.; Ghatty, S.; Reddy, A.R. De novo transcriptome analysis of an imminent biofuel crop, Camelina sativa L. using Illumina GAIIX sequencing platform and identification of SSR markers. Plant Mol. Biol. 2014, 84, 159–171. [Google Scholar] [CrossRef]
- Suresh, S.; Park, J.-H.; Cho, G.-T.; Lee, H.-S.; Baek, H.-J.; Lee, S.-Y.; Chung, J.-W. Development and molecular characterization of 55 novel polymorphic cDNA-SSR markers in faba bean (Vicia faba L.) using 454 pyrosequencing. Molecules 2013, 18, 1844–1856. [Google Scholar] [CrossRef]
- Fu, N.; Wang, Q.; Shen, H.-L. De novo assembly, gene annotation and marker development using illumina paired-end transcriptome sequences in celery (Apium graveolens L.). PLoS One 2013, 8, e57686. [Google Scholar]
- Cloutier, S.; Miranda, E.; Ward, K.; Radovanovic, N.; Reimer, E.; Walichnowski, A.; Datla, R.; Rowland, G.; Duguid, S.; Ragupathy, R. Simple sequence repeat marker development from bacterial artificial chromosome end sequences and expressed sequence tags of flax (Linum usitatissimum L.). Theor. Appl. Genet. 2012, 125, 685–694. [Google Scholar] [CrossRef]
- Eujayl, I.; Sorrells, M.; Baum, M.; Wolters, P.; Powell, W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet. 2002, 104, 399–407. [Google Scholar]
- Jena, S.N.; Srivastava, A.; Rai, K.M.; Ranjan, A.; Singh, S.K.; Nisar, T.; Srivastava, M.; Bag, S.K.; Mantri, S.; Asif, M.H. Development and characterization of genomic and expressed SSRs for levant cotton (Gossypium herbaceum L.). Theor. Appl. Genet. 2012, 124, 565–576. [Google Scholar]
- Pashley, C.H.; Ellis, J.R.; McCauley, D.E.; Burke, J.M. EST databases as a source for molecular markers: Lessons from Helianthus. J. Hered. 2006, 97, 381–388. [Google Scholar]
- Laurent, V.; Devaux, P.; Thiel, T.; Viard, F.; Mielordt, S.; Touzet, P.; Quillet, M. Comparative effectiveness of sugar beet microsatellite markers isolated from genomic libraries and GenBank ESTs to map the sugar beet genome. Theor. Appl. Genet. 2007, 115, 793–805. [Google Scholar] [CrossRef]
- Liewlaksaneeyanawin, C.; Ritland, C.E.; El-Kassaby, Y.A.; Ritland, K. Single-copy, species-transferable microsatellite markers developed from loblolly pine ESTs. Theor. Appl. Genet. 2004, 109, 361–369. [Google Scholar]
- Ramu, P.; Billot, C.; Rami, J.; Senthilvel, S.; Upadhyaya, H.; Reddy, L.A.; Hash, C. Assessment of genetic diversity in the sorghum reference set using EST-SSR markers. Theor. Appl. Genet. 2013, 126, 2051–2064. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Li, J.; Zhao, Y.; Korban, S.S.; Han, Y. Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Mol. Biol. Rep. 2012, 30, 539–546. [Google Scholar] [CrossRef]
- Wang, H.F.; Zong, X.X.; Guan, J.P.; Yang, T.; Sun, X.L.; Ma, Y.; Redden, R. Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theor. Appl. Genet. 2012, 124, 789–797. [Google Scholar] [CrossRef]
- De Andrés, M.; Benito, A.; Pérez-Rivera, G.; Ocete, R.; Lopez, M.; Gaforio, L.; Muñoz, G.; Cabello, F.; Martínez Zapater, J.M.; Arroyo-García, R. Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol. Ecol. 2012, 21, 800–816. [Google Scholar] [CrossRef]
- Mostafa, N.; Omar, H.; Tan, S.G.; Napis, S. Studies on the genetic variation of the green unicellular alga Haematococcus pluvialis (Chlorophyceae) obtained from different geographical locations using ISSR and RAPD molecular marker. Molecules 2011, 16, 2599–2608. [Google Scholar] [CrossRef]
- Belaj, A.; Muñoz-Diez, C.; Baldoni, L.; Porceddu, A.; Barranco, D.; Satovic, Z. Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Ann. Bot. 2007, 100, 449–458. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, C.Q.; Li, D.Z. Low genetic diversity and high genetic differentiation in the critically endangered Omphalogramma souliei (Primulaceae): Implications for its conservation. J. Syst. Evol. 2009, 47, 103–109. [Google Scholar] [CrossRef]
- Ozkan, H.; Kafkas, S.; Ozer, M.S.; Brandolini, A. Genetic relationships among South-East Turkey wild barley populations and sampling strategies of Hordeum spontaneum. Theor. Appl. Genet. 2005, 112, 12–20. [Google Scholar] [CrossRef]
- Jia, X.-D.; Wang, T.; Zhai, M.; Li, Y.-R.; Guo, Z.-R. Genetic diversity and identification of chinese-grown pecan using ISSR and SSR markers. Molecules 2011, 16, 10078–10092. [Google Scholar] [CrossRef]
- Simko, I.; Eujayl, I.; van Hintum, T.J. Empirical evaluation of DArT, SNP, and SSR marker-systems for genotyping, clustering, and assigning sugar beet hybrid varieties into populations. Plant Sci. 2012, 184, 54–62. [Google Scholar] [CrossRef]
- Caruso, M.; Federici, C.T.; Roose, M.L. EST–SSR markers for asparagus genetic diversity evaluation and cultivar identification. Mol. Breed. 2008, 21, 195–204. [Google Scholar]
- Kabelka, E.; Franchino, B.; Francis, D. Two loci from Lycopersicon hirsutum LA407 confer resistance to strains of Clavibacter michiganensis subsp. michiganensis. Phytopathology 2002, 92, 504–510. [Google Scholar] [CrossRef]
- Primer3 (version 0.4.0). Available online: http://bioinfo.ut.ee/primer3-0.4.0/primer3/ (accessed on 10 February 2014).
- Weir, B.S. Genetic Data Analysis. In Methods for Discrete Population Genetic Data; Sinauer Associates: Sunderland, MA, USA, 1990. [Google Scholar]
- Yeh, F.; Yang, R.-C.; Boyle, T. PopGene Version 131: Microsoft Window-Based Freeware for Population Genetic Analysis. Available online: http://www.ualberta.ca/~fyeh/popgene.pdf (accessed on 7 February 2014).
- Levene, H. On a matching problem arising in genetics. Ann. Math. Stat. 1949, 20, 91–94. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar]
- Rohlf, F.J. Ntsys-Pc Numerical Taxonomy and Multivariate Analysis System Version 2.1, Exeter Software; Setauket: New York, NY, USA, 2000. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Agapow, P.M.; Burt, A. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 2001, 1, 101–102. [Google Scholar] [CrossRef]
- Sample Availability: All samples are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fu, N.; Wang, P.-Y.; Liu, X.-D.; Shen, H.-l. Use of EST-SSR Markers for Evaluating Genetic Diversity and Fingerprinting Celery (Apium graveolens L.) Cultivars. Molecules 2014, 19, 1939-1955. https://doi.org/10.3390/molecules19021939
Fu N, Wang P-Y, Liu X-D, Shen H-l. Use of EST-SSR Markers for Evaluating Genetic Diversity and Fingerprinting Celery (Apium graveolens L.) Cultivars. Molecules. 2014; 19(2):1939-1955. https://doi.org/10.3390/molecules19021939
Chicago/Turabian StyleFu, Nan, Ping-Yong Wang, Xiao-Dan Liu, and Huo-lin Shen. 2014. "Use of EST-SSR Markers for Evaluating Genetic Diversity and Fingerprinting Celery (Apium graveolens L.) Cultivars" Molecules 19, no. 2: 1939-1955. https://doi.org/10.3390/molecules19021939




