Anticancer Agents Targeted to Sirtuins
Abstract
:1. Introduction
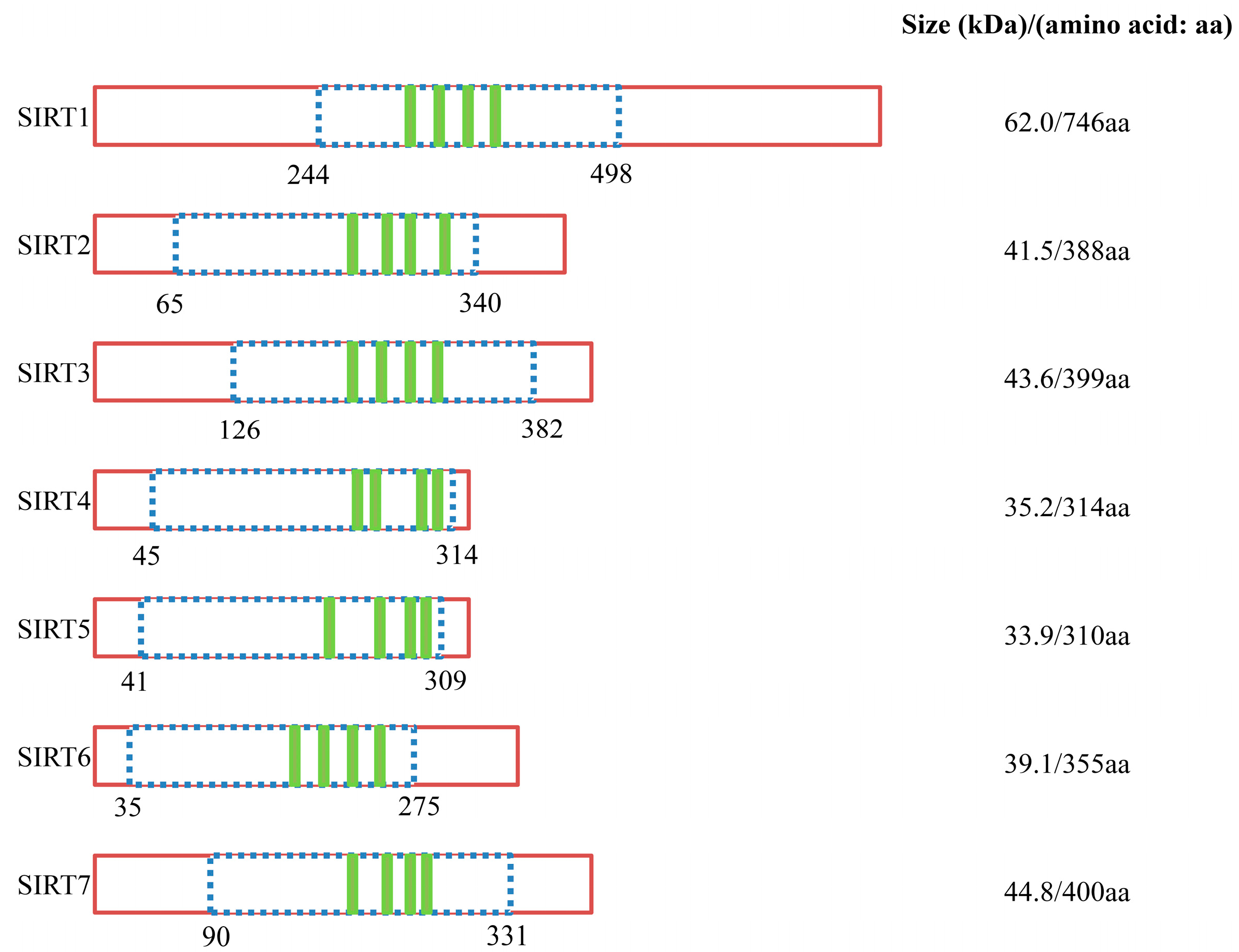
| Class | Sirtuin | Enzyme Activity | Location | Main Substrates | High Expression | Functions |
|---|---|---|---|---|---|---|
| I | SIRT1 | Deacetylase | Nuclear/cytoplasmic | Histone H1K26, H3K9, H3K18, H4K16, p53, FOXO, Rb, Ku70, p300, NF-κB, PGC-1α, PPARγ, UCP2, Acetyl-CoA synthetase 1, MnSOD, Smad7, MyoD, Per2, SUV39H1 | Brain, skeletal muscle, heart, kidney and rterus | Cell survival, lifespan regulation, metabolism regulation, inflamation, oxidative stress response |
| I | SIRT2 | Deacetylase | Nuclear/cytoplasmic | α-tubulin, FOXO, Histone H4K16 | Brain | Cell cycle regulation, nervous system development |
| I | SIRT3 | Deacetylase | Mitochondrial | Histone H3K9, H4K16, Acetyl-CoA synthetase 2, Ku70, FOXO3a, MnSOD | Brain, heat, liver, kidney and brown adipose tissue | Regulation of mitochondrial metabolism, fatty acid oxidation, ATP-production |
| II | SIRT4 | ADP-ribosyl-transferase | Mitochondrial | Glutamate dehydrogenase | Pancreatic b-cellls, brain, liver, kidney and heart | Regulation of mitochondrial metabolism, insulin secretion |
| III | SIRT5 | Deacetylase/Demalonylase/Desuccinylase | Mitochondrial | Carbamoyl phospate synthetase 1, Cytochrome c, Carbamoyl phosphate synthetase 1 | Brain, testis, heart, muscle and lymphoblast | Apoptosis, regulation of protein-protein interactiond and protein stability, urea cycle |
| IV | SIRT6 | Deacetylase/ADP-ribosyl-transferase | Nuclear | Histone H3K9, H3K56, TNF-α, HIF1α | Brain, muscle, heart, ovary and bone cells (absent in bone marrow) | Genome stability (base excision repair), DNA rapair |
| IV | SIRT7 | Deacetylase | Nuclear | Histone H3K18, p53 | Peripheral blood cells, CD33+ myeloid bone marrow precuseor cells | Regulation of rRNA transcription, cell cycle regulation |
2. Sirtuins in Cancer
3. Sirtuins and Cell Death
4. Sirtuin Inhibitors
| Inhibitors | Structure | MW | Target (IC50) |
|---|---|---|---|
| Splitomicin |  | 198.22 | Sir2 (60 μM) |
| EX-527 |  | 248.71 | SIRT1 (98 nM) |
| Sirtinol |  | 394.47 | SIRT1 (131 μM), |
| SIRT2 (49 μM) | |||
| Cambinol |  | 360.43 | SIRT1 (56 μM), |
| SIRT2 (59 μM) | |||
| Salermide |  | 394.5 | SIRT1 (76.2 μM), |
| SIRT2 (45 μM) | |||
| Tenovin-6 |  | 454.6 | SIRT1 (21 μM), |
| SIRT2 (10 μM), | |||
| SIRT3 (67 μM) | |||
| Suramin |  | 1429.17 | SIRT1 (297 nM), |
| SIRT2 (1.2 μM), | |||
| SIRT5 (22 μM) | |||
| AGK2 |  | 434.27 | SIRT2 (3.5 μM) |
5. Sirtuin Activators
6. Conclusions
Acknowledgments
Author Contributions
Abbreviations
| ATG | autophagy-related protein |
| ATL | adult T-cell leukemia-lymphoma |
| BRCA1 | breast cancer susceptibility gene 1 |
| FOXO | forkhead-box |
| H3K18Ac | H3 acetyl lysine 18 |
| HIC1 | hypermethylated in cancer 1 |
| HTLV-1 | human T-cell leukemia virus |
| IC50 | 50% inhibitory concentration |
| NAD | nicotinamide adenine dinucleotide |
| NF-κB | nuclear factor-κB |
| Rb | retinoblastoma |
| RIP3 | receptor-interacting protein kinase 3 |
| Sir2 | Silencing information regulator 2 |
| TGF-β | transforming growth factor-β |
| TNF-α | tumor necrosis factor-α |
Conflicts of Interest
References
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein sir2 is an nad-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.; Sutton, A.; Tafrov, S.T.; Heller, R.C.; Stebbins, J.; Pillus, L.; Sternglanz, R. The silencing protein sir2 and its homologs are nad-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 2000, 97, 5807–5811. [Google Scholar] [CrossRef] [PubMed]
- Tanny, J.C.; Dowd, G.J.; Huang, J.; Hilz, H.; Moazed, D. An enzymatic activity in the yeast sir2 protein that is essential for gene silencing. Cell 1999, 99, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Rine, J.; Herskowitz, I. Four genes responsible for a position effect on expression from hml and hmr in saccharomyces cerevisiae. Genetics 1987, 116, 9–22. [Google Scholar] [PubMed]
- Bordone, L.; Guarente, L. Calorie restriction, sirt1 and metabolism: Understanding longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.Y.; Miller, C.; Bitterman, K.J.; Wall, N.R.; Hekking, B.; Kessler, B.; Howitz, K.T.; Gorospe, M.; de Cabo, R.; Sinclair, D.A. Calorie restriction promotes mammalian cell survival by inducing the sirt1 deacetylase. Science 2004, 305, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Sanders, B.D.; Jackson, B.; Marmorstein, R. Structural basis for sirtuin function: What we know and what we don’t. Biochim. Biophys. Acta 2010, 1804, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Garva, R.; Krstic-Demonacos, M.; Demonacos, C. Sirtuins: Molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription. J. Biomed. Biotechnol. 2011, 2011, 368276. [Google Scholar] [CrossRef] [PubMed]
- Lavu, S.; Boss, O.; Elliott, P.J.; Lambert, P.D. Sirtuins—Novel therapeutic targets to treat age-associated diseases. Nat. Rev. Drug Discov. 2008, 7, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 is a nad-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.; Chen, C.Y.; Ho, K.K.; Di Fruscia, P.; Myatt, S.S.; Coombes, R.C.; Fuchter, M.J.; Hsiao, C.D.; Lam, E.W. Sirt inhibitors induce cell death and p53 acetylation through targeting both sirt1 and sirt2. Mol. Cancer Ther. 2010, 9, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Alcain, F.J.; Villalba, J.M. Sirtuin activators. Expert Opin. Ther. Pat. 2009, 19, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Alcain, F.J.; Villalba, J.M. Sirtuin inhibitors. Expert Opin. Ther. Pat. 2009, 19, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.H.; Sengupta, K.; Li, C.; Kim, H.S.; Cao, L.; Xiao, C.; Kim, S.; Xu, X.; Zheng, Y.; Chilton, B.; et al. Impaired DNA damage response, genome instability, and tumorigenesis in sirt1 mutant mice. Cancer Cell 2008, 14, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Kim, S.J.; Sandal, B.; Lee, S.M.; Gao, B.; Zhang, D.D.; Fang, D. The type III histone deacetylase Sirt1 protein suppresses p300-mediated histone H3 lysine 56 acetylation at Bclaf1 promoter to inhibit T cell activation. J. Biol. Chem. 2011, 286, 16967–16975. [Google Scholar] [CrossRef] [PubMed]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappab-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, R.; Bres, V.; Ng, R.W.; Coudart, M.P.; El Messaoudi, S.; Sardet, C.; Jin, D.Y.; Emiliani, S.; Benkirane, M. Post-activation turn-off of NF-kappa b-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 2003, 278, 2758–2766. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.H.; Zheng, Y.; Kim, H.S.; Xu, X.; Cao, L.; Luhasen, T.; Lee, M.H.; Xiao, C.; Vassilopoulos, A.; Chen, W.; et al. Interplay among BRCA1, Sirt1, and survivin during BRCA1-associated tumorigenesis. Mol. Cell 2008, 32, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Chou, P.M.; Zheng, X.; Mirkin, B.L.; Rebbaa, A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005, 65, 10183–10187. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, P.Y.; Marshall, G.M. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009, 69, 1702–1705. [Google Scholar] [CrossRef] [PubMed]
- Kozako, T.; Aikawa, A.; Shoji, T.; Fujimoto, T.; Yoshimitsu, M.; Shirasawa, S.; Tanaka, H.; Honda, S.; Shimeno, H.; Arima, N.; et al. High expression of the longevity gene product SIRT1 and apoptosis induction by sirtinol in adult T-cell leukemia cells. Int. J. Cancer 2012, 131, 2044–2055. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; Dessain, S.K.; Ng Eaton, E.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.M.; Wang, Z.Y.; Sun, D.C.; Lin, R.X.; Wang, S.Q. Sirt1 interacts with p73 and suppresses p73-dependent transcriptional activity. J. Cell. Physiol. 2007, 210, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Wang, D.H.; Yen, R.C.; Luo, J.; Gu, W.; Baylin, S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005, 123, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Tseng, R.C.; Lee, C.C.; Hsu, H.S.; Tzao, C.; Wang, Y.C. Distinct HIC1-SIRT1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia 2009, 11, 763–770. [Google Scholar] [PubMed]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by sir2alpha promotes cell survival under stress. Cell 2001, 107, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Wales, M.M.; Biel, M.A.; el Deiry, W.; Nelkin, B.D.; Issa, J.P.; Cavenee, W.K.; Kuerbitz, S.J.; Baylin, S.B. P53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat. Med. 1995, 1, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Vassilopoulos, A.; Wang, R.H.; Lahusen, T.; Xiao, Z.; Xu, X.; Li, C.; Veenstra, T.D.; Li, B.; Yu, H.; et al. Sirt2 maintains genome integrity and suppresses tumorigenesis through regulating apc/c activity. Cancer Cell 2011, 20, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Hiratsuka, M.; Osaki, M.; Oshimura, M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle 2007, 6, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, N.; Takami, Y.; Kimura, M.; Tachiiri, S.; Ishiai, M.; Nakayama, T.; Takata, M. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells 2005, 10, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Harting, K.; Knoll, B. SIRT2-mediated protein deacetylation: An emerging key regulator in brain physiology and pathology. Eur. J. Cell Biol. 2010, 89, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, M.; Inoue, T.; Toda, T.; Kimura, N.; Shirayoshi, Y.; Kamitani, H.; Watanabe, T.; Ohama, E.; Tahimic, C.G.; Kurimasa, A.; et al. Proteomics-based identification of differentially expressed genes in human gliomas: Down-regulation of SIRT2 gene. Biochem. Biophys. Res. Commun. 2003, 309, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strathearn, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.C.; McLean, P.J.; et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 2007, 317, 516–519. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Nie, H.; Hong, Y.; Sheng, C.; Xia, W.; Ying, W. Sirt2 activity is required for the survival of c6 glioma cells. Biochem. Biophys. Res. Commun. 2012, 417, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Matsumori, H.; Nakayama, Y.; Osaki, M.; Kojima, H.; Kurimasa, A.; Ito, H.; Mori, S.; Katoh, M.; Oshimura, M.; et al. Sirt2 down-regulation in hela can induce p53 accumulation via p38 mapk activation-dependent p300 decrease, eventually leading to apoptosis. Genes Cells 2011, 16, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; et al. Sirt7 links h3k18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487, 114–118. [Google Scholar] [PubMed]
- Manuyakorn, A.; Paulus, R.; Farrell, J.; Dawson, N.A.; Tze, S.; Cheung-Lau, G.; Hines, O.J.; Reber, H.; Seligson, D.B.; Horvath, S.; et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: Results from rtog 9704. J. Clin. Oncol. 2010, 28, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.B.; Horvath, S.; McBrian, M.A.; Mah, V.; Yu, H.; Tze, S.; Wang, Q.; Chia, D.; Goodglick, L.; Kurdistani, S.K. Global levels of histone modifications predict prognosis in different cancers. Am. J. Pathol. 2009, 174, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, N.; Kikuchi, J.; Koyama, D.; Wada, T.; Mori, S.; Nakamura, Y.; Furukawa, Y. Alkylating agents induce histone h3k18 hyperacetylation and potentiate hdac inhibitor-mediated global histone acetylation and cytotoxicity in mantle cell lymphoma. Blood Cancer J. 2013, 3, e169. [Google Scholar] [CrossRef] [PubMed]
- Stunkel, W.; Peh, B.K.; Tan, Y.C.; Nayagam, V.M.; Wang, X.; Salto-Tellez, M.; Ni, B.; Entzeroth, M.; Wood, J. Function of the sirt1 protein deacetylase in cancer. Biotechnol. J. 2007, 2, 1360–1368. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, C.A.; Khanim, F.L.; Hayden, R.; Bunce, C.M.; White, D.A.; Drayson, M.T.; Craddock, C.; Turner, B.M. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia 2005, 19, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Kozako, T. New strategy of adult t-cell leukemia treatment targeted for anti-tumor immunity and a longevity gene-encoded protein. Yakugaku Zasshi 2011, 131, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Ouaissi, M.; Sielezneff, I.; Silvestre, R.; Sastre, B.; Bernard, J.P.; Lafontaine, J.S.; Payan, M.J.; Dahan, L.; Pirro, N.; Seitz, J.F.; et al. High histone deacetylase 7 (hdac7) expression is significantly associated with adenocarcinomas of the pancreas. Ann. Surg. Oncol. 2008, 15, 2318–2328. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zuo, Y.; Feng, Y.; Zhang, M. Sirt5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumor Biol. 2014, 35, 10699–10705. [Google Scholar] [CrossRef]
- Inoue, T.; Hiratsuka, M.; Osaki, M.; Yamada, H.; Kishimoto, I.; Yamaguchi, S.; Nakano, S.; Katoh, M.; Ito, H.; Oshimura, M. Sirt2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene 2007, 26, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Lennerz, V.; Fatho, M.; Gentilini, C.; Frye, R.A.; Lifke, A.; Ferel, D.; Wolfel, C.; Huber, C.; Wolfel, T. The response of autologous t cells to a human melanoma is dominated by mutated neoantigens. Proc. Natl. Acad. Sci. USA 2005, 102, 16013–16018. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, C.; Zwaans, B.M.; Silberman, D.M.; Gymrek, M.; Goren, A.; Zhong, L.; Ram, O.; Truelove, J.; Guimaraes, A.R.; Toiber, D.; et al. The histone deacetylase sirt6 is a tumor suppressor that controls cancer metabolism. Cell 2012, 151, 1185–1199. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Patel, K.; Muldoon-Jacobs, K.; Bisht, K.S.; Aykin-Burns, N.; Pennington, J.D.; van der Meer, R.; Nguyen, P.; Savage, J.; Owens, K.M.; et al. Sirt3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010, 17, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.M.; Fraga, M.F. Aging and cancer: Are sirtuins the link? Future Oncol. 2010, 6, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Vakhrusheva, O.; Smolka, C.; Gajawada, P.; Kostin, S.; Boettger, T.; Kubin, T.; Braun, T.; Bober, E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 2008, 102, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.; Ichim, G.; Green, D.R. Die another way—Non-apoptotic mechanisms of cell death. J. Cell Sci. 2014, 127, 2135–2144. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.; Green, D.R. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Shao, S.H.; Xu, Z.X.; Hennessy, B.; Ding, Z.; Larrea, M.; Kondo, S.; Dumont, D.J.; Gutterman, J.U.; Walker, C.L.; et al. The energy sensing lkb1-ampk pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007, 9, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Bonapace, L.; Bornhauser, B.C.; Schmitz, M.; Cario, G.; Ziegler, U.; Niggli, F.K.; Schafer, B.W.; Schrappe, M.; Stanulla, M.; Bourquin, J.P. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J. Clin. Investig. 2010, 120, 1310–1323. [Google Scholar] [CrossRef] [PubMed]
- Farkas, T.; Daugaard, M.; Jaattela, M. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J. Biol. Chem. 2011, 286, 38904–38912. [Google Scholar] [CrossRef] [PubMed]
- Osborn, S.L.; Diehl, G.; Han, S.J.; Xue, L.; Kurd, N.; Hsieh, K.; Cado, D.; Robey, E.A.; Winoto, A. Fas-associated death domain (FADD) is a negative regulator of T-cell receptor-mediated necroptosis. Proc. Natl. Acad. Sci. USA 2010, 107, 13034–13039. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.; Green, D.R. Caspase-independent cell death: Leaving the set without the final cut. Oncogene 2008, 27, 6452–6461. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A role for the NAD-dependent deacetylase SIRT1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Yang, H.; Kong, Q.; Li, J.; Lee, S.M.; Gao, B.; Dong, H.; Wei, J.; Song, J.; Zhang, D.D.; et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 2012, 46, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of foxo transcription factors by the sirt1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Ford, J.; Jiang, M.; Milner, J. Cancer-specific functions of sirt1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005, 65, 10457–10463. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.L.; Gu, W. P53 activation: A case against sir. Cancer Cell 2008, 13, 377–378. [Google Scholar] [CrossRef] [PubMed]
- Langley, E.; Pearson, M.; Faretta, M.; Bauer, U.M.; Frye, R.A.; Minucci, S.; Pelicci, P.G.; Kouzarides, T. Human sir2 deacetylates p53 and antagonizes pml/p53-induced cellular senescence. EMBO J. 2002, 21, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. P53 mutations in human cancers. Science 1991, 253, 49–53. [Google Scholar] [CrossRef]
- Yi, J.; Luo, J. Sirt1 and p53, effect on cancer, senescence and beyond. Biochim. Biophys. Acta 2010, 1804, 1684–1689. [Google Scholar] [CrossRef] [PubMed]
- Han, M.K.; Song, E.K.; Guo, Y.; Ou, X.; Mantel, C.; Broxmeyer, H.E. Sirt1 regulates apoptosis and nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell 2008, 2, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Dimova, D.K.; Dyson, N.J. The E2F transcriptional network: Old acquaintances with new faces. Oncogene 2005, 24, 2810–2826. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Weber, J.D. Deacetylation of the retinoblastoma tumour suppressor protein by sirt1. Biochem. J. 2007, 407, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Brunet, A. Foxo transcription factors at the interface between longevity and tumor suppression. Oncogene 2005, 24, 7410–7425. [Google Scholar] [CrossRef] [PubMed]
- Barreyro, F.J.; Kobayashi, S.; Bronk, S.F.; Werneburg, N.W.; Malhi, H.; Gores, G.J. Transcriptional regulation of bim by foxo3a mediates hepatocyte lipoapoptosis. J. Biol. Chem. 2007, 282, 27141–27154. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Tindall, D.J. Foxos, cancer and regulation of apoptosis. Oncogene 2008, 27, 2312–2319. [Google Scholar] [CrossRef]
- Kume, S.; Haneda, M.; Kanasaki, K.; Sugimoto, T.; Araki, S.; Isshiki, K.; Isono, M.; Uzu, T.; Guarente, L.; Kashiwagi, A.; et al. Sirt1 inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells via smad7 deacetylation. J. Biol. Chem. 2007, 282, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.F.; Mostoslavsky, R.; Lombard, D.B.; Pang, W.W.; Saito, S.; Franco, S.; Kaushal, D.; Cheng, H.L.; Fischer, M.R.; Stokes, N.; et al. Mammalian sirt1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005, 2, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.J.; Milner, J. Sirt3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle 2007, 6, 2669–2677. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nguyen, M.; Qin, F.X.; Tong, Q. SIRT2 deacetylates FOXO3A in response to oxidative stress and caloric restriction. Aging Cell 2007, 6, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Bell, B.D.; Leverrier, S.; Weist, B.M.; Newton, R.H.; Arechiga, A.F.; Luhrs, K.A.; Morrissette, N.S.; Walsh, C.M. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16677–16682. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, L.; Yang, J.; Zhang, P.; Ma, K.; Zhou, J.; Liao, W.; Zhu, W.G. Anti-neoplastic activity of the cytosolic foxo1 results from autophagic cell death. Autophagy 2010, 6, 988–990. [Google Scholar] [CrossRef] [PubMed]
- Villalba, J.M.; Alcain, F.J. Sirtuin activators and inhibitors. Biofactors 2012, 38, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Bedalov, A.; Gatbonton, T.; Irvine, W.P.; Gottschling, D.E.; Simon, J.A. Identification of a small molecule inhibitor of sir2p. Proc. Natl. Acad. Sci. USA 2001, 98, 15113–15118. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, R.C.; Uchiechowska, U.; Meier, R.; Hruby, H.; Valkov, V.; Verdin, E.; Sippl, W.; Jung, M. Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J. Med. Chem. 2008, 51, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.M.; Pasupuleti, R.; Xu, L.; McDonagh, T.; Curtis, R.; DiStefano, P.S.; Huber, L.J. Inhibition of sirt1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 2006, 26, 28–38. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Chao, E.D.; Blackwell, H.E.; Moazed, D.; Schreiber, S.L. Identification of a class of small molecule inhibitors of the sirtuin family of nad-dependent deacetylases by phenotypic screening. J. Biol. Chem. 2001, 276, 38837–38843. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Tokunaga, E.; Chang, K.; Hikasa, M.; Iijima, K.; Eto, M.; Kozaki, K.; Akishita, M.; Ouchi, Y.; Kaneki, M. Sirt1 inhibitor, sirtinol, induces senescence-like growth arrest with attenuated ras-mapk signaling in human cancer cells. Oncogene 2006, 25, 176–185. [Google Scholar] [PubMed]
- Kojima, K.; Ohhashi, R.; Fujita, Y.; Hamada, N.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. A role for sirt1 in cell growth and chemoresistance in prostate cancer pc3 and du145 cells. Biochem. Biophys. Res. Commun. 2008, 373, 423–428. [Google Scholar] [CrossRef]
- Wang, J.; Kim, T.H.; Ahn, M.Y.; Lee, J.; Jung, J.H.; Choi, W.S.; Lee, B.M.; Yoon, K.S.; Yoon, S.; Kim, H.S. Sirtinol, a class III HDAC inhibitor, induces apoptotic and autophagic cell death in MCF-7 human breast cancer cells. Int. J. Oncol. 2012, 41, 1101–1109. [Google Scholar] [PubMed]
- Takeda-Watanabe, A.; Kitada, M.; Kanasaki, K.; Koya, D. SIRT1 inactivation induces inflammation through the dysregulation of autophagy in human THP-1 cells. Biochem. Biophys. Res. Commun. 2012, 427, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Heltweg, B.; Gatbonton, T.; Schuler, A.D.; Posakony, J.; Li, H.; Goehle, S.; Kollipara, R.; Depinho, R.A.; Gu, Y.; Simon, J.A.; et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006, 66, 4368–4377. [Google Scholar] [CrossRef] [PubMed]
- Lara, E.; Mai, A.; Calvanese, V.; Altucci, L.; Lopez-Nieva, P.; Martinez-Chantar, M.L.; Varela-Rey, M.; Rotili, D.; Nebbioso, A.; Ropero, S.; et al. Salermide, a sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 2009, 28, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Lain, S.; Hollick, J.J.; Campbell, J.; Staples, O.D.; Higgins, M.; Aoubala, M.; McCarthy, A.; Appleyard, V.; Murray, K.E.; Baker, L.; et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 2008, 13, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Hirai, S.; Endo, S.; Saito, R.; Hirose, M.; Ueno, T.; Suzuki, H.; Yamato, K.; Abei, M.; Hyodo, I. Antitumor effects of a sirtuin inhibitor, tenovin-6, against gastric cancer cells via death receptor 5 up-regulation. PLoS One 2014, 9, e102831. [Google Scholar] [CrossRef] [PubMed]
- MacCallum, S.F.; Groves, M.J.; James, J.; Murray, K.; Appleyard, V.; Prescott, A.R.; Drbal, A.A.; Nicolaou, A.; Cunningham, J.; Haydock, S.; et al. Dysregulation of autophagy in chronic lymphocytic leukemia with the small-molecule sirtuin inhibitor tenovin-6. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.; Min, J.; Antoshenko, T.; Wang, C.L.; Allali-Hassani, A.; Dong, A.; Loppnau, P.; Vedadi, M.; Bochkarev, A.; Sternglanz, R.; et al. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure 2007, 15, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Imai, K.; Imai, E.; Iida, S.; Ueda, R.; Tsumoto, H.; Nakagawa, H.; Miyata, N. Design, synthesis, enzyme inhibition, and tumor cell growth inhibition of 2-anilinobenzamide derivatives as SIRT1 inhibitors. Bioorg. Med. Chem. 2009, 17, 5900–5905. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Khan, M.N.; Sawada, H.; Imai, E.; Itoh, Y.; Yamatsuta, K.; Tokuda, N.; Takeuchi, J.; Seko, T.; Nakagawa, H.; et al. Design, synthesis, and biological activity of a novel series of human sirtuin-2-selective inhibitors. J. Med. Chem. 2012, 55, 5760–5773. [Google Scholar] [CrossRef] [PubMed]
- Nebbioso, A.; Pereira, R.; Khanwalkar, H.; Matarese, F.; Garcia-Rodriguez, J.; Miceli, M.; Logie, C.; Kedinger, V.; Ferrara, F.; Stunnenberg, H.G.; et al. Death receptor pathway activation and increase of ROS production by the triple epigenetic inhibitor UVI5008. Mol. Cancer Ther. 2011, 10, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Kim, M.J.; Lee, S.H.; Lee, J.W.; Bae, J.H.; Kim, D.W.; Dao, T.T.; Oh, W.K.; Kang, C.D.; Kim, S.H. Amurensin g, a novel sirt1 inhibitor, sensitizes trail-resistant human leukemic k562 cells to trail-induced apoptosis. Biochem. Pharmacol. 2012, 84, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Kalle, A.M.; Mallika, A.; Badiger, J.; Alinakhi; Talukdar, P. Sachchidanand. Inhibition of sirt1 by a small molecule induces apoptosis in breast cancer cells. Biochem. Biophys. Res. Commun. 2010, 401, 13–19. [Google Scholar] [CrossRef]
- Pirrie, L.; McCarthy, A.R.; Major, L.L.; Morkunaite, V.; Zubriene, A.; Matulis, D.; Lain, S.; Lebl, T.; Westwood, N.J. Discovery and validation of SIRT2 inhibitors based on tenovin-6: Use of a 1H-NMR method to assess deacetylase activity. Molecules 2012, 17, 12206–12224. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, G.A.; Zhang, K.; McBrian, M.A.; Grunstein, M.; Kurdistani, S.K.; Berk, A.J. Adenovirus small e1a alters global patterns of histone modification. Science 2008, 321, 1084–1085. [Google Scholar] [CrossRef] [PubMed]
- Eskandarian, H.A.; Impens, F.; Nahori, M.A.; Soubigou, G.; Coppee, J.Y.; Cossart, P.; Hamon, M.A. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 2013, 341. [Google Scholar] [CrossRef]
- Suzuki, Y.; Ito, S.; Sasaki, R.; Asahi, M.; Ishida, Y. Resveratrol suppresses cell proliferation via inhibition of STAT3 phosphorylation and McL-1 and cIAP-2 expression in HTLV-1-infected T cells. Leuk. Res. 2013, 37, 1674–1679. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.E.; Patel, B.B.; Wang, J.; Arabshahi, A.; Eltoum, I.A.; Lamartiniere, C.A. Resveratrol suppresses prostate cancer progression in transgenic mice. Carcinogenesis 2007, 28, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Whitsett, T.; Carpenter, M.; Lamartiniere, C.A. Resveratrol, but not EGCG, in the diet suppresses DMBA-induced mammary cancer in rats. J. Carcinog. 2006, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.H.; Afaq, F.; Ahmad, N. Prevention of ultraviolet-b radiation damage by resveratrol in mouse skin is mediated via modulation in survivin. Photochem. Photobiol. 2005, 81, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Marino, G.; Bennetzen, M.V.; Eisenberg, T.; Megalou, E.; Schroeder, S.; Cabrera, S.; Benit, P.; Rustin, P.; Criollo, A.; et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011, 192, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, X.; Zhu, J.X.; Xie, W.; Le, W.; Fan, Z.; Jankovic, J.; Pan, T. Resveratrol-activated ampk/sirt1/autophagy in cellular models of parkinson’s disease. Neurosignals 2011, 19, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Ray, D.; Lekli, I.; Bak, I.; Tosaki, A.; Das, D.K. Effects of longevinex (modified resveratrol) on cardioprotection and its mechanisms of action. Can. J. Physiol. Pharmacol. 2010, 88, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Timmers, S.; Auwerx, J.; Schrauwen, P. The journey of resveratrol from yeast to human. Aging (Albany N. Y.) 2012, 4, 146–158. [Google Scholar]
- Milne, J.C.; Lambert, P.D.; Schenk, S.; Carney, D.P.; Smith, J.J.; Gagne, D.J.; Jin, L.; Boss, O.; Perni, R.B.; Vu, C.B.; et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 2007, 450, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Bandi, M.; Singh, A.V.; Ray, A.; Raje, N.; Richardson, P.; Anderson, K.C. Preclinical evaluation of a novel SIRT1 modulator SRT1720 in multiple myeloma cells. Br. J. Haematol. 2011, 155, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Pacholec, M.; Bleasdale, J.E.; Chrunyk, B.; Cunningham, D.; Flynn, D.; Garofalo, R.S.; Griffith, D.; Griffor, M.; Loulakis, P.; Pabst, B.; et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010, 285, 8340–8351. [Google Scholar] [CrossRef] [PubMed]
- Borra, M.T.; Smith, B.C.; Denu, J.M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005, 280, 17187–17195. [Google Scholar] [CrossRef] [PubMed]
- Beher, D.; Wu, J.; Cumine, S.; Kim, K.W.; Lu, S.C.; Atangan, L.; Wang, M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 2009, 74, 619–624. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozako, T.; Suzuki, T.; Yoshimitsu, M.; Arima, N.; Honda, S.-i.; Soeda, S. Anticancer Agents Targeted to Sirtuins. Molecules 2014, 19, 20295-20313. https://doi.org/10.3390/molecules191220295
Kozako T, Suzuki T, Yoshimitsu M, Arima N, Honda S-i, Soeda S. Anticancer Agents Targeted to Sirtuins. Molecules. 2014; 19(12):20295-20313. https://doi.org/10.3390/molecules191220295
Chicago/Turabian StyleKozako, Tomohiro, Takayoshi Suzuki, Makoto Yoshimitsu, Naomichi Arima, Shin-ichiro Honda, and Shinji Soeda. 2014. "Anticancer Agents Targeted to Sirtuins" Molecules 19, no. 12: 20295-20313. https://doi.org/10.3390/molecules191220295
APA StyleKozako, T., Suzuki, T., Yoshimitsu, M., Arima, N., Honda, S.-i., & Soeda, S. (2014). Anticancer Agents Targeted to Sirtuins. Molecules, 19(12), 20295-20313. https://doi.org/10.3390/molecules191220295





