Structures of Some Novel α-Glucosyl Diterpene Glycosides from the Glycosylation of Steviol Glycosides
Abstract
:1. Introduction
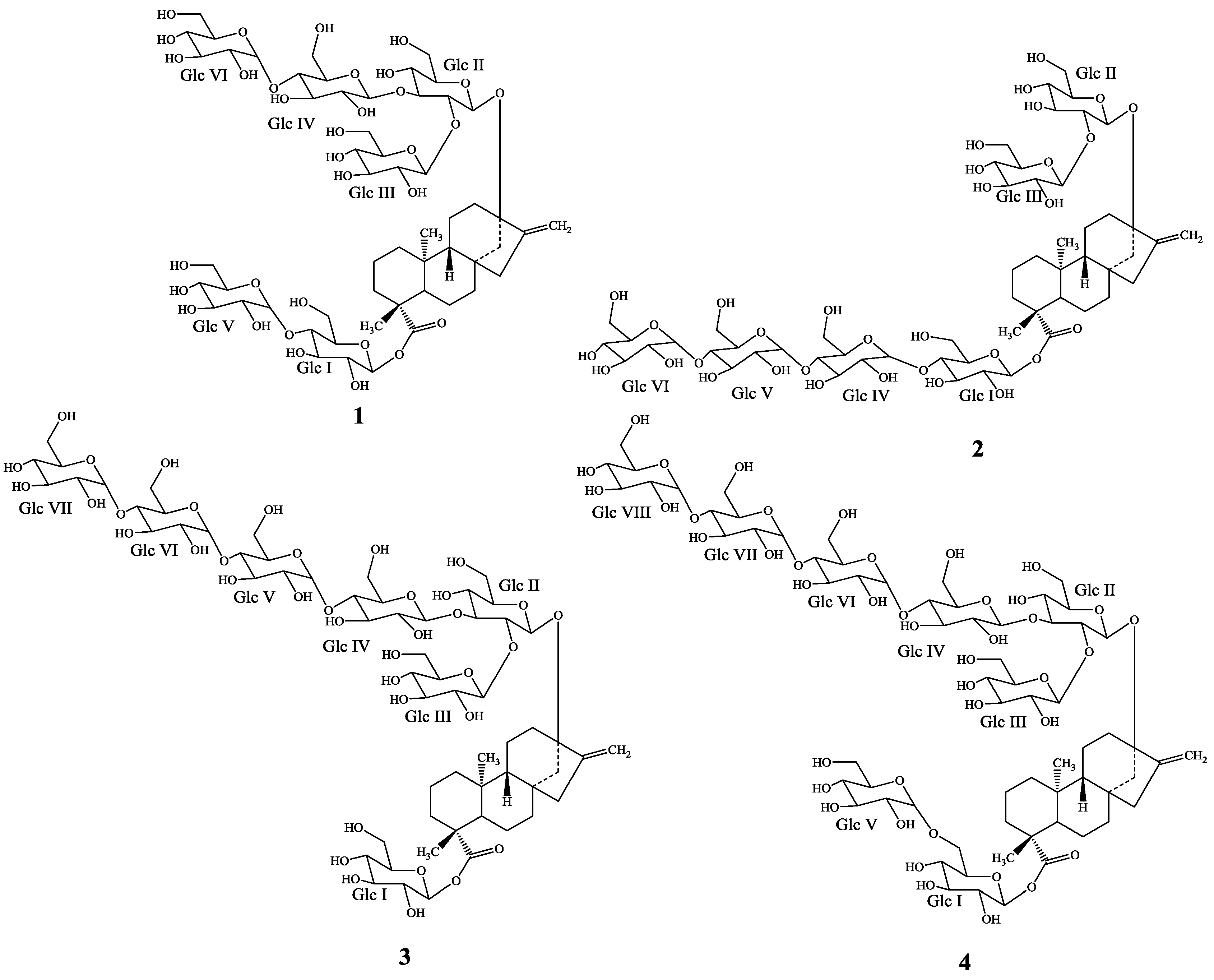
2. Results and Discussion
| 1H | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 0.76 t (12.1)1.74 m | 0.77 t (12.5)1.74 m | 0.77 t (11.5)1.76 d (12.1) | 0.76 t (10.8)1.75 m |
| 2 | 1.46 m2.15 m | 1.45 m2.16 m | 1.47 m2.18 m | 1.45 d (12.3)2.17 m |
| 3 | 1.04 m2.34 m | 1.05 m2.34 m | 1.05 m2.35 m | 1.04 m2.33 d (11.7) |
| 4 | - | - | - | - |
| 5 | 1.05 d (12.5) | 1.06 d (12.0) | 1.06 d (12.2) | 1.05 d (11.4) |
| 6 | 1.90 m2.33 m | 1.90 m2.38 m | 1.91 m2.42 m | 1.91 m2.33 d (11.5) |
| 7 | 1.32 m1.38 m | 1.35 m | 1.33 m1.37 m | 1.33 m1.37 m |
| 8 | - | - | - | - |
| 9 | 0.90 m | 0.91 m | 0.90 d (6.9) | 0.90 d (6.9) |
| 10 | - | - | - | - |
| 11 | 1.67 m1.70 m | 1.66 m | 1.68 m1.70 m | 1.67 m1.70 m |
| 12 | 1.91 m2.24 m | 1.89 m2.24 m | 1.93 m2.25 m | 1.90 m2.23 m |
| 13 | - | - | - | - |
| 14 | 1.79 m2.63 d (11.7) | 1.79 d (11.2)2.70 d (11.1) | 1.83 d (11.7)2.64 d (11.4) | 1.79 d (11.0)2.63 d (11.0) |
| 15 | 2.04 d (17.2)2.11 d (17.2) | 2.05 d (17.3)2.11 d (17.3) | 2.04 d (17.2)2.11 d (17.2) | 2.03 d (17.7)2.10 d (17.7) |
| 16 | - | - | - | - |
| 17 | 5.05 s5.68 s | 5.10 s5.73 s | 5.05 s5.68 s | 5.06 s5.69 s |
| 18 | 1.26 s | 1.27 s | 1.27 s | 1.26 s |
| 19 | - | - | - | - |
| 20 | 1.23 s | 1.25 s | 1.27 s | 1.23 s |
| 1' | 5.98 d (8.4) | 6.08 d (8.3) | 6.07 d (8.4) | 5.98 d (8.2) |
| 2' | 4.08 t (8.4) | 4.13 m | 4.12 m | 4.08 t (8.6) |
| 3' | 4.28 m | 4.40 m | 4.23 m | 4.28 m |
| 4' | 4.31 m | 4.33 m | 4.24 m | 4.31 m |
| 5' | 3.74 m | 3.95 m | 3.98 | 3.72 m |
| 6' | 4.32 m | 4.28 m4.43 | ||
| 1'' | 5.07 d (8.1) | 5.13 d (7.7) | 5.06 d (7.6) | 5.05 d (7.6) |
| 2'' | 4.36 m | 4.21 | 4.39 m | 4.38 m |
| 3'' | 4.30 m | 4.32 | 4.31 m | 4.36 m |
| 4'' | 3.89 m | 4.03 | 3.89 t (8.6) | 3.91 t (8.5) |
| 5'' | 3.77 m | 3.88 | 3.80 t (7.6) | 3.77 t (7.6) |
| 6'' | 4.09 m4.30 m | 4.214.31 | 4.10 m4.43 m | 4.11 m |
| 1''' | 5.58 d (7.8) | 5.32 d (7.6) | 5.60 d (7.8) | 5.60 d (7.8) |
| 2''' | 4.13 m | 4.16 | 4.16 m | 4.14 m |
| 3''' | 4.27 m | 4.25 | 4.29 m | 4.28 m |
| 4''' | 4.18 m | 4.28 | 4.19 m | 4.19 m |
| 5''' | 3.96 m | 3.96 | 3.97 m | 3.97 m |
| 6''' | 4.33 m4.55 m | 4.394.53 | 4.34 m4.54 m | 4.33 m4.54 m |
| 1'''' | 5.32 d (7.9) | 5.80 d (3.7) | 5.45 d (7.8) | 5.47 d (7.6) |
| 2'''' | 3.97 m | 4.11 | 4.00 m | 4.00 m |
| 3'''' | 4.20 m | 4.57 | 4.32 m | 4.31 m |
| 4'''' | 4.14 m | 4.14 | 4.13 m | 4.13 m |
| 5'''' | 3.83 m | 4.03 m | 4.02 m | |
| 6'''' | 4.30 m4.52 m | 4.29 m4.54 m | 4.314.54 m | |
| 1''''' | 5.81 d (3.8) | 5.75 d (3.8) | 5.74 d (3.1) | 5.87 d (3.4) |
| 2''''' | 4.16 m | 4.13 | 4.13 m | 4.14 m |
| 3''''' | 4.55 m | 4.61 | 4.59 m | 4.54 m |
| 4''''' | 4.12 m | 4.21 | 4.15 m | 4.12 m |
| 5''''' | 4.49 m | 4.34 | 4.32 m | 4.49 m |
| 6''''' | ||||
| 1'''''' | 5.87 d (3.7) | 5.89 d (3.7) | 5.78 d (3.1) | 5.73 d (3.2) |
| 2'''''' | 4.14 m | 4.19 | 4.14 m | 4.12 m |
| 3'''''' | 4.55 m | 4.57 | 4.62 t (9.3) | 4.59 m |
| 4'''''' | 4.12 m | 4.14 | 4.21 m | 4.16 m |
| 5'''''' | 4.49 m | NC | 4.34 m | 4.32 m |
| 6'''''' | NC | NC | NC | NC |
| 1''''''' | 5.90 d (3.2) | 5.78 d (3.2) | ||
| 2''''''' | 4.19 m | 4.14 m | ||
| 3''''''' | 4.58 m | 4.62 t (9.2) | ||
| 4''''''' | 4.16 m | 4.21 m | ||
| 5''''''' | 4.54 m | 4.33 m | ||
| 6''''''' | NC | NC | ||
| 1'''''''' | 5.90 d (3.2) | |||
| 2'''''''' | 4.19 m | |||
| 3'''''''' | 4.56 m | |||
| 4'''''''' | 4.12 m | |||
| 5'''''''' | NC | |||
| 6'''''''' |
| 13C | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 40.7 | 40.8 | 40.8 | 40.8 |
| 2 | 19.5 | 19.5 | 19.5 | 19.7 |
| 3 | 38.4 | 38.4 | 38.4 | 38.4 |
| 4 | 44.2 | 44.2 | 44.2 | 44.3 |
| 5 | 57.3 | 57.3 | 57.4 | 57.4 |
| 6 | 22.2 | 22.2 | 22.1 | 22.1 |
| 7 | 41.6 | 41.7 | 41.8 | 41.7 |
| 8 | - | - | - | - |
| 9 | 54.1 | 54.0 | 54.1 | 54.1 |
| 10 | 39.5 | 39.4 | 39.5 | 39.5 |
| 11 | 20.5 | 20.7 | 20.6 | 20.5 |
| 12 | 37.3 | 36.7 | 37.2 | 37.3 |
| 13 | 86.7 | 86.6 | 86.9 | |
| 14 | 44.7 | 44.8 | 44.6 | 44.8 |
| 15 | 47.8 | 47.7 | 47.8 | 47.9 |
| 16 | - | - | - | - |
| 17 | 105.1 | 105.2 | 105.0 | 105.2 |
| 18 | 28.5 | 28.5 | 28.5 | 28.6 |
| 19 | 177.3 | 177.7 | 177.9 | 177.6 |
| 20 | 15.8 | 15.8 | 15.7 | 15.7 |
| 1' | 95.4 | 95.6 | 95.7 | 95.4 |
| 2' | 73.3 | 73.3 | 73.9 | 73.3 |
| 3' | 77.9 | 78.0 | 78.7 | 78.0 |
| 4' | 80.1 | 80.7 | 70.6 | 80.1 |
| 5' | 77.4 | ND | 79.0 | 77.3 |
| 6' | ND | ND | ND | ND |
| 1'' | 97.8 | 97.8 | 97.8 | 97.7 |
| 2'' | 80.5 | 83.5 | 80.6 | 80.5 |
| 3'' | 87.0 | 78.1 | 86.9 | 87.1 |
| 4'' | 70.1 | 71.8 | 70.2 | 70.1 |
| 5'' | 77.3 | 77.7 | 77.3 | 77.2 |
| 6'' | 62.2 | 62.4 | ND | ND |
| 1''' | 104.2 | 106.1 | 104.3 | 104.2 |
| 2''' | 76.1 | 76.5 | 76.1 | 76.0 |
| 3''' | 78.1 | 78.0 | 78.2 | 78.0 |
| 4''' | 71.8 | 71.5 | 71.8 | 71.8 |
| 5''' | 78.4 | ND | 78.5 | 78.4 |
| 6''' | 63.0 | ND | ND | ND |
| 1'''' | 104.2 | 102.7 | 104.2 | 104.1 |
| 2'''' | 74.5 | 73.5 | 74.5 | 74.6 |
| 3'''' | 77.8 | 74.7 | 77.6 | 77.8 |
| 4'''' | 81.3 | 82.0 | 81.9 | 81.9 |
| 5'''' | 76.8 | ND | 76.9 | 76.9 |
| 6'''' | 62.6 | ND | ND | ND |
| 1''''' | 102.9 | 103.0 | 102.9 | 102.7 |
| 2''''' | 74.0 | 73.5 | 73.4 | 73.9 |
| 3''''' | 75.0 | 74.8 | 74.7 | 75.0 |
| 4''''' | 71.6 | 81.4 | 81.9 | 71.7 |
| 5''''' | 75.1 | ND | 73.3 | 75.1 |
| 6''''' | ND | ND | ND | |
| 1'''''' | 102.7 | 102.9 | 102.9 | 102.8 |
| 2'''''' | 74.0 | 74.1 | 73.2 | 73.4 |
| 3'''''' | 75.0 | 75.4 | 74.7 | 74.8 |
| 4'''''' | 71.6 | 71.7 | 81.5 | 81.9 |
| 5'''''' | 75.1 | ND | 73.3 | 73.4 |
| 6'''''' | ND | ND | ND | |
| 1''''''' | 103.0 | 102.9 | ||
| 2''''''' | 74.0 | 73.4 | ||
| 3''''''' | 74.9 | 74.8 | ||
| 4''''''' | 71.8 | 81.5 | ||
| 5''''''' | 75.1 | 73.4 | ||
| 6''''''' | ND | ND | ||
| 1'''''''' | 103.0 | |||
| 2'''''''' | 74.1 | |||
| 3'''''''' | 74.9 | |||
| 4'''''''' | 71.7 | |||
| 5'''''''' | ND | |||
| 6'''''''' | ND |

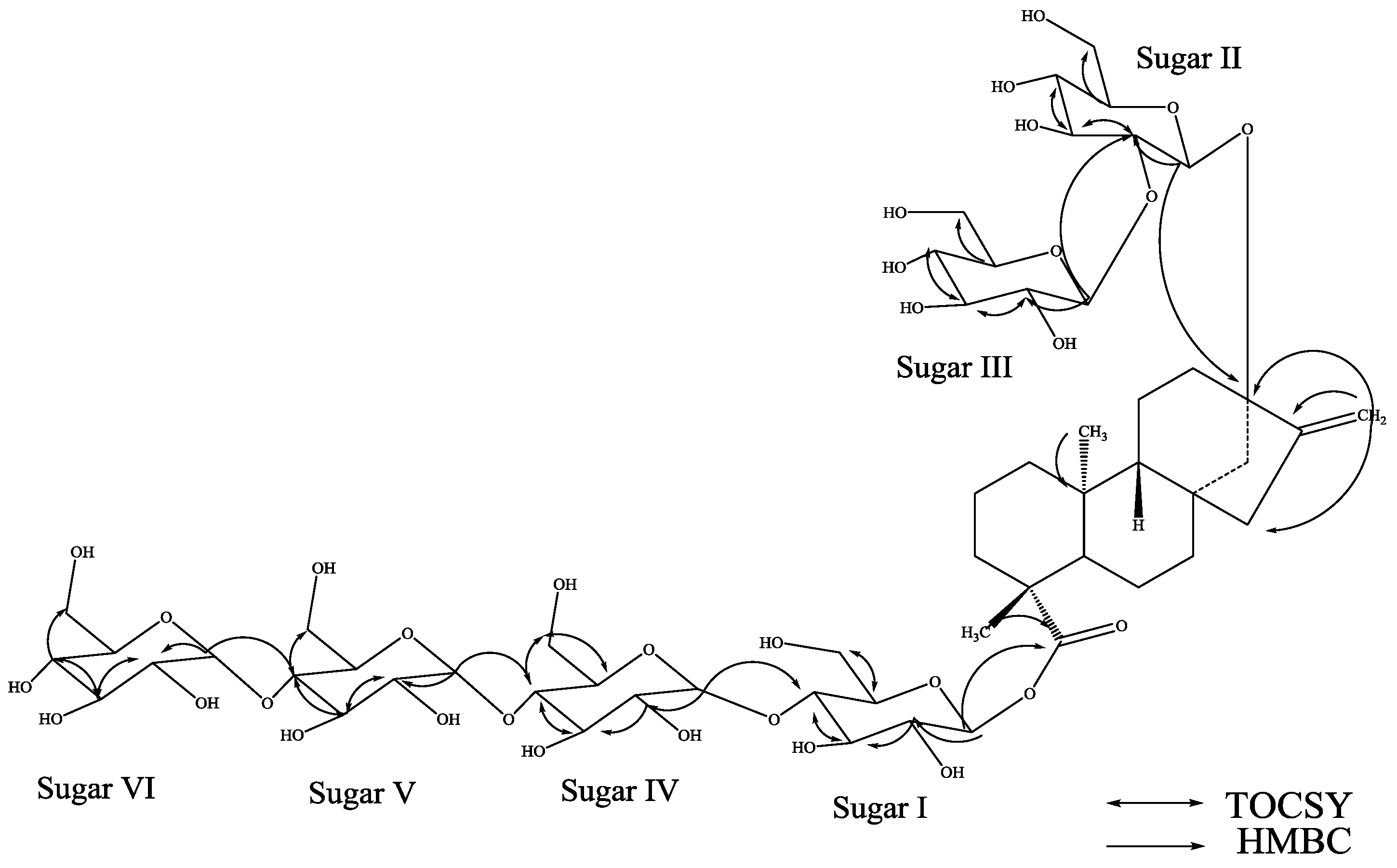
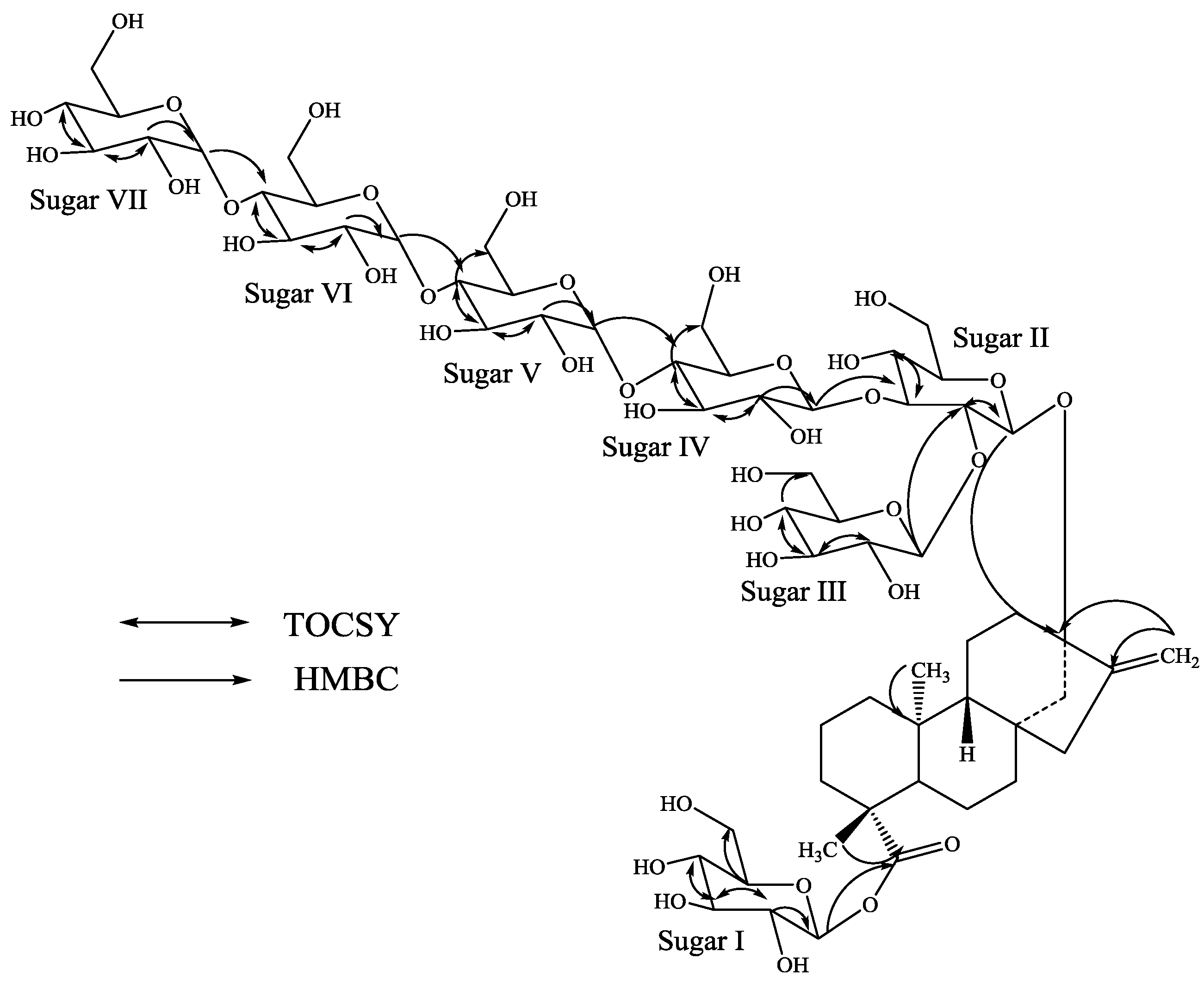

3. Experimental Section
3.1. General
3.2. Plant Material
3.3. Isolation
| HPLC Method | Time (min) | % of Mobile Phase A | % of Mobile Phase B |
|---|---|---|---|
| Method 1 | 0.0 | 75 | 25 |
| 8.5 | 75 | 25 | |
| 10.0 | 71 | 29 | |
| 16.5 | 70 | 30 | |
| 18.5 | 66 | 34 | |
| 24.5 | 66 | 34 | |
| 25.0 | 0 | 100 | |
| 30.0 | 0 | 100 | |
| Method 2 | 0.0 | 72 | 28 |
| 60.0 | 72 | 28 | |
| Method 3 | 0.0 | 20 | 80 |
| 100.0 | 20 | 80 |
General Procedure for Acid Hydrolysis and Determination of Sugar Configuration in 1–4
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lewis, W.H. Early uses of Stevia rebaudiana (Asteraceae) leaves as a sweetener in Paraguay. Econ. Bot. 1992, 46, 336–337. [Google Scholar]
- Ibrahim, M.A.; Rodenburg, D.L.; Alves, K.; Fronczek, F.R.; McChesney, J.D.; Wu, C.; Nettles, B.J.; Venkataraman, S.K.; Jacksch, F. Minor diterpene glycosides from the leaves of Stevia rebaudiana. J. Nat. Prod. 2014, 77, 1231–1235. [Google Scholar]
- Mosettig, E.; Nes, W.R. Stevioside. II. The structure of the aglucon. J. Org. Chem. 1955, 20, 884–899. [Google Scholar]
- Mosettig, E.; Beglinger, U.; Dolder, F.; Lichiti, H.; Quitt, P.; Waters, J.A. The absolute configuration of steviol and isosteviol. J. Am. Chem. Soc. 1963, 85, 2305–2309. [Google Scholar]
- Brandle, J.E.; Starrratt, A.N.; Gijen, M. Stevia rebaudiana: Its agricultural, biological and chemical properties. Can. J. Plant Sci. 1998, 78, 527–536. [Google Scholar]
- Wayne, E.S.; Lin, L. NMR studies of the conformation of the natural sweetener rebaudioside A. Carbohydr. Res. 2009, 344, 2533–2538. [Google Scholar]
- Chaturvedula, V.S.P.; Mani, U.; Prakash, I. Structures of the novel α-glucosyl linked diterpene glycosides from Stevia rebaudiana. Carbohydr. Res. 2011, 346, 2034–2038. [Google Scholar]
- Prakash, I.; Chaturvedula, V.S.P.; Markosyan, A. Isolation, characterization and sensory evaluation of a hexa β-d-glucopyranosyl diterpene from Stevia rebaudiana. Nat. Prod. Commun. 2013, 8, 1523–1526. [Google Scholar]
- Prakash, I.; Chaturvedula, V.S.P. Additional minor diterpene glycosides from Stevia rebaudiana bertoni. Molecules 2013, 18, 13510–13519. [Google Scholar]
- Chaturvedula, V.S.P.; Mani, U.; Prakash, I. Diterpene glycosides from Stevia rebaudiana. Molecules 2011, 16, 3552–3562. [Google Scholar]
- Chaturvedula, V.S.P.; Prakash, I. A new diterpenoid glycoside from Stevia rebaudiana. Molecules 2011, 16, 2937–2943. [Google Scholar]
- Chaturvedula, V.S.P.; Prakash, I. Structures of the novel diterpene glycosides from Stevia rebaudiana. Carbohydr. Res. 2011, 346, 1057–1060. [Google Scholar]
- Bedir, E.; Toyang, N.J.; Khan, I.A.; Walker, L.A.; Clark, A.M. A new dammarane type triterpene glycoside from Polyscias fulva. J. Nat. Prod. 2001, 64, 95–97. [Google Scholar]
- Chaturvedula, V.S.P.; Schilling, J.K.; Miller, J.S.; Andriantsiferana, R.; Rasamison, V.E.; Kingston, D.G.I. New cytotoxic oleanane saponis from the infructescences of Polyscias amplifolia from the Madagascar rainforest. Planta Med. 2003, 69, 440–444. [Google Scholar]
- Huan, V.D.; Yamamura, S.; Ohtani, K.; Kasai, R.; Yamasaki, K.; Nham, N.T. Oleanane saponins from Polyscias fructicosa. Phytochemistry 1998, 47, 451–457. [Google Scholar]
- Tanaka, T.; Nakashima, T.; Ueda, T.; Tomii, K.; Kouno, I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. 2007, 55, 899–901. [Google Scholar]
- Abelyan, V.; Markosyan, A.; Abelyan, L. Sweetner and use. U.S. Patent Application No. 2010/0166679, 1 July 2010. [Google Scholar]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakash, I.; Chaturvedula, V.S.P. Structures of Some Novel α-Glucosyl Diterpene Glycosides from the Glycosylation of Steviol Glycosides. Molecules 2014, 19, 20280-20294. https://doi.org/10.3390/molecules191220280
Prakash I, Chaturvedula VSP. Structures of Some Novel α-Glucosyl Diterpene Glycosides from the Glycosylation of Steviol Glycosides. Molecules. 2014; 19(12):20280-20294. https://doi.org/10.3390/molecules191220280
Chicago/Turabian StylePrakash, Indra, and Venkata Sai Prakash Chaturvedula. 2014. "Structures of Some Novel α-Glucosyl Diterpene Glycosides from the Glycosylation of Steviol Glycosides" Molecules 19, no. 12: 20280-20294. https://doi.org/10.3390/molecules191220280




