Analysis on the Interaction Domain of VirG and Apyrase by Pull-Down Assay
Abstract
:1. Introduction
2. Results and Discussion
2.1. Apyrase Interacts with VirG
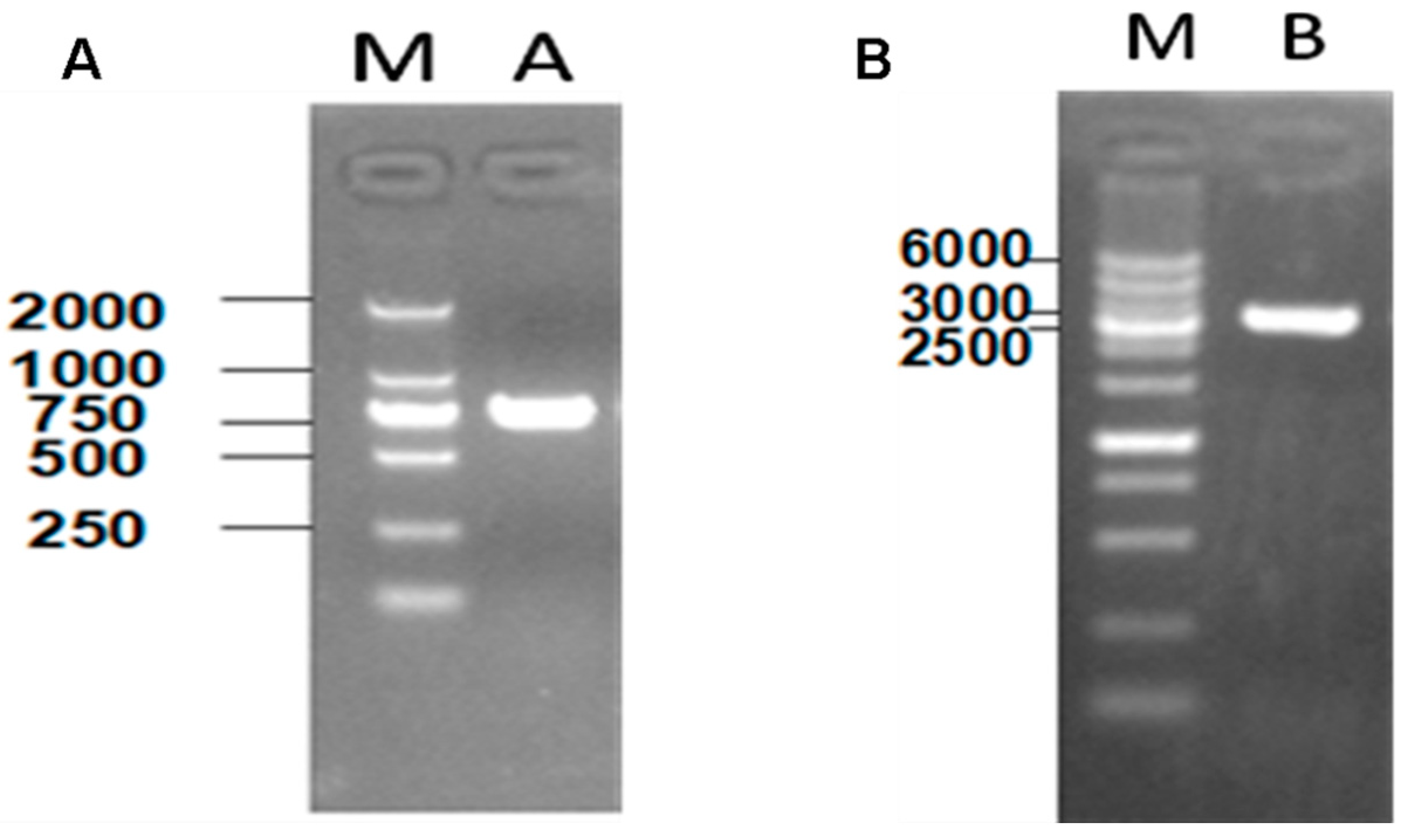
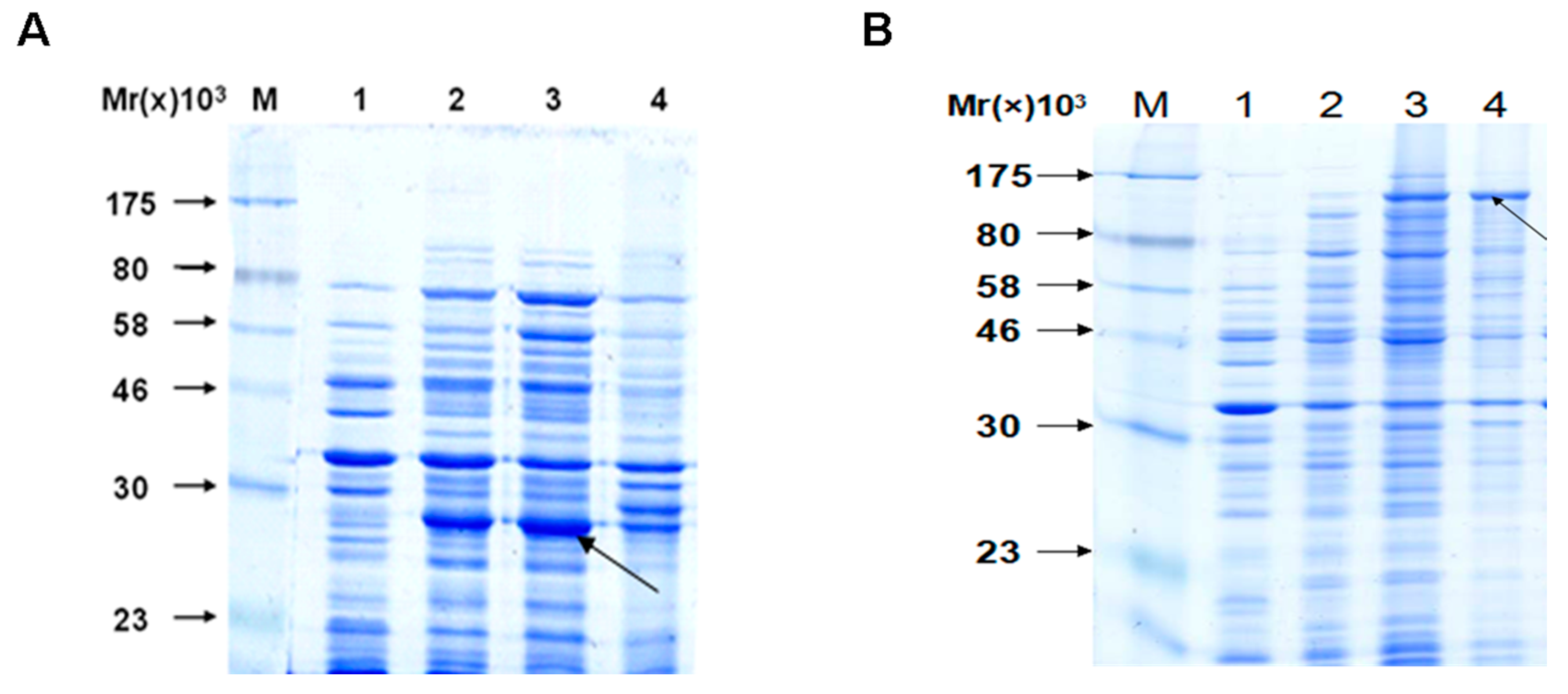
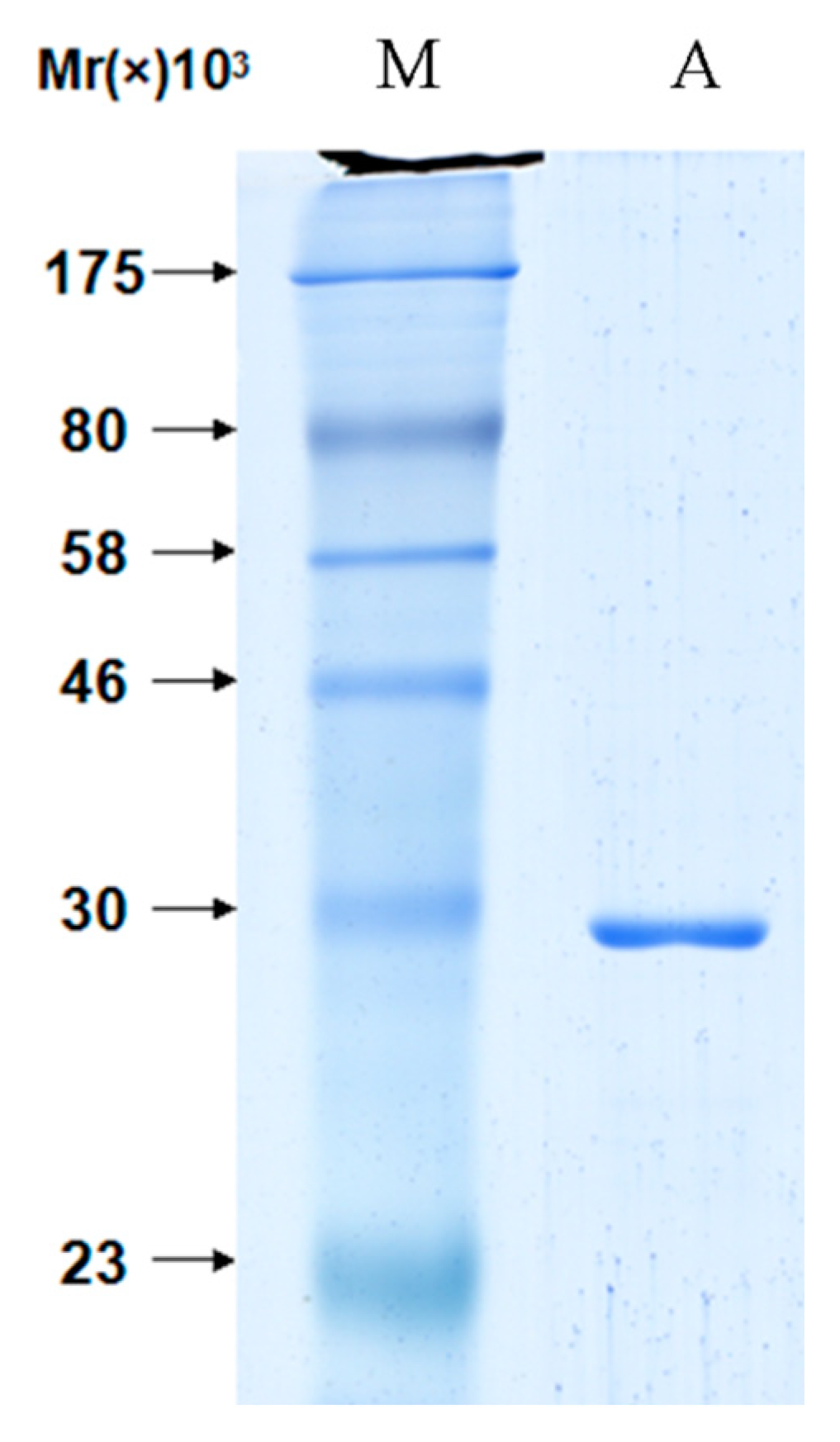
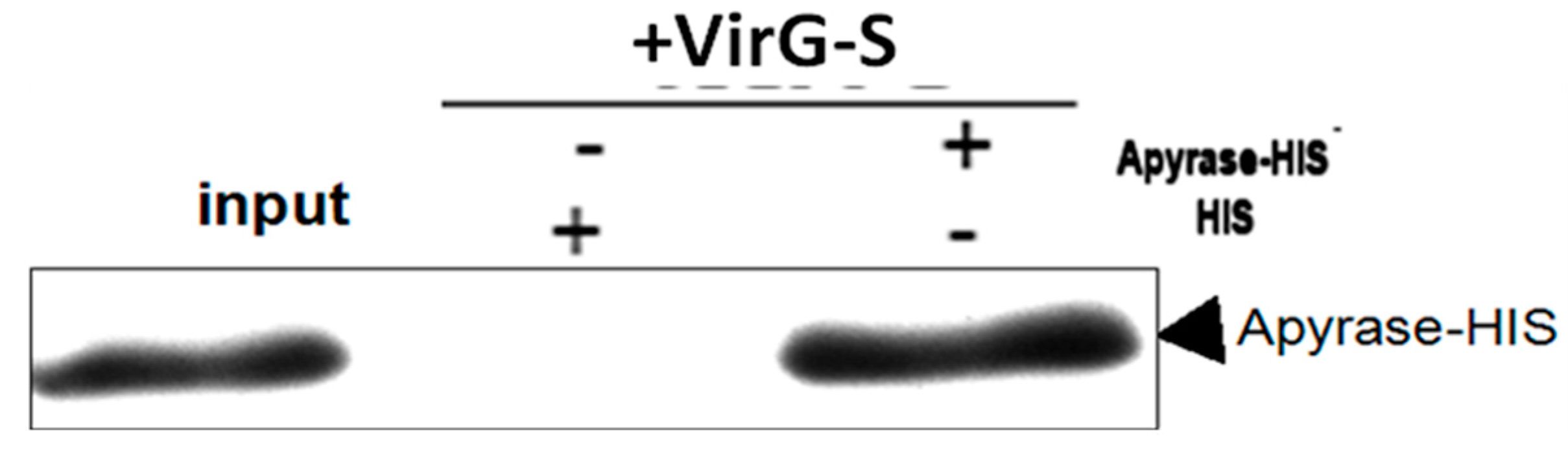
2.2. The Interaction between Apyrase and Residues 507–758 of the α-Domain Was Identified
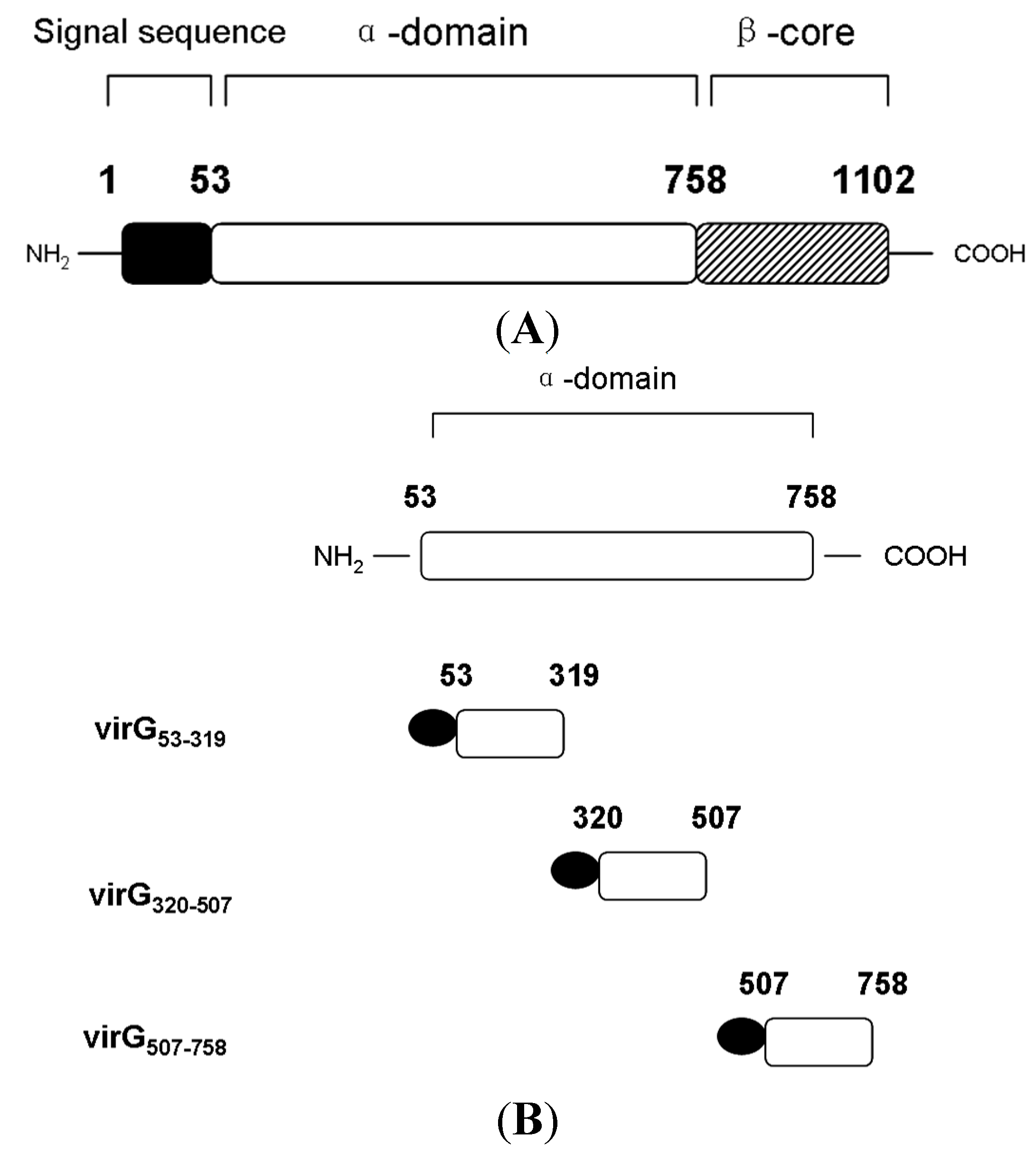
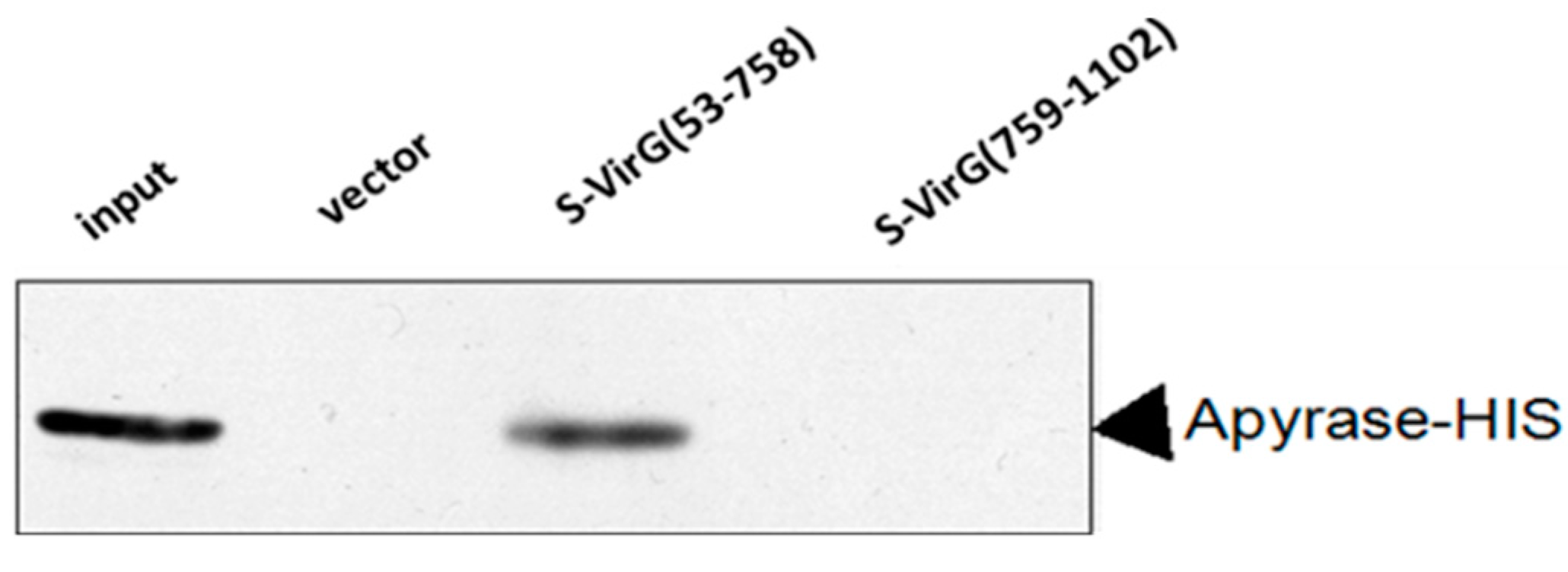
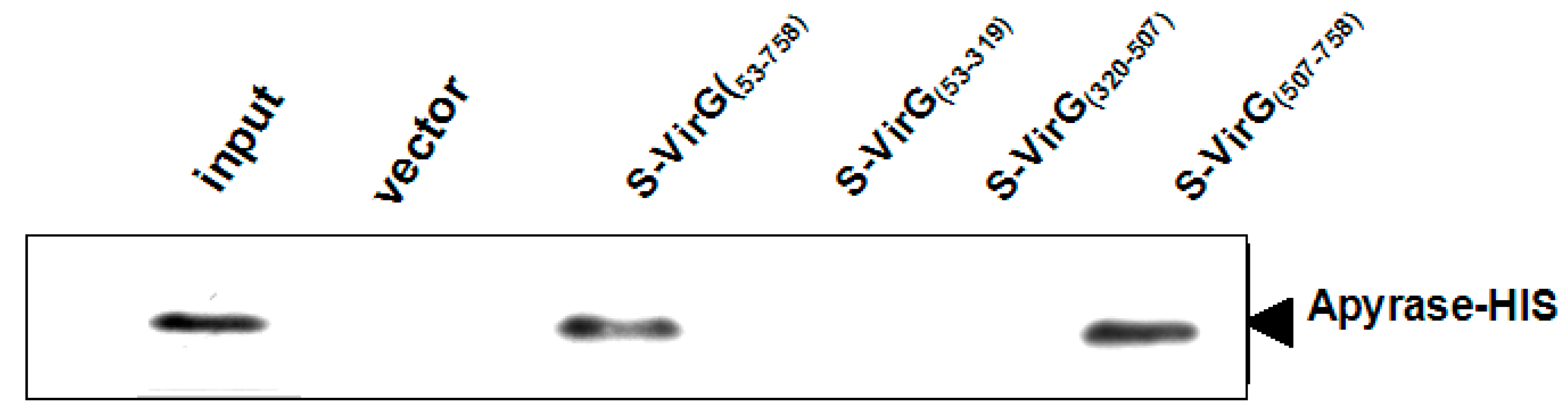
2.3. The Function of VirG (IcsA) Was Influenced in ΔphoN2
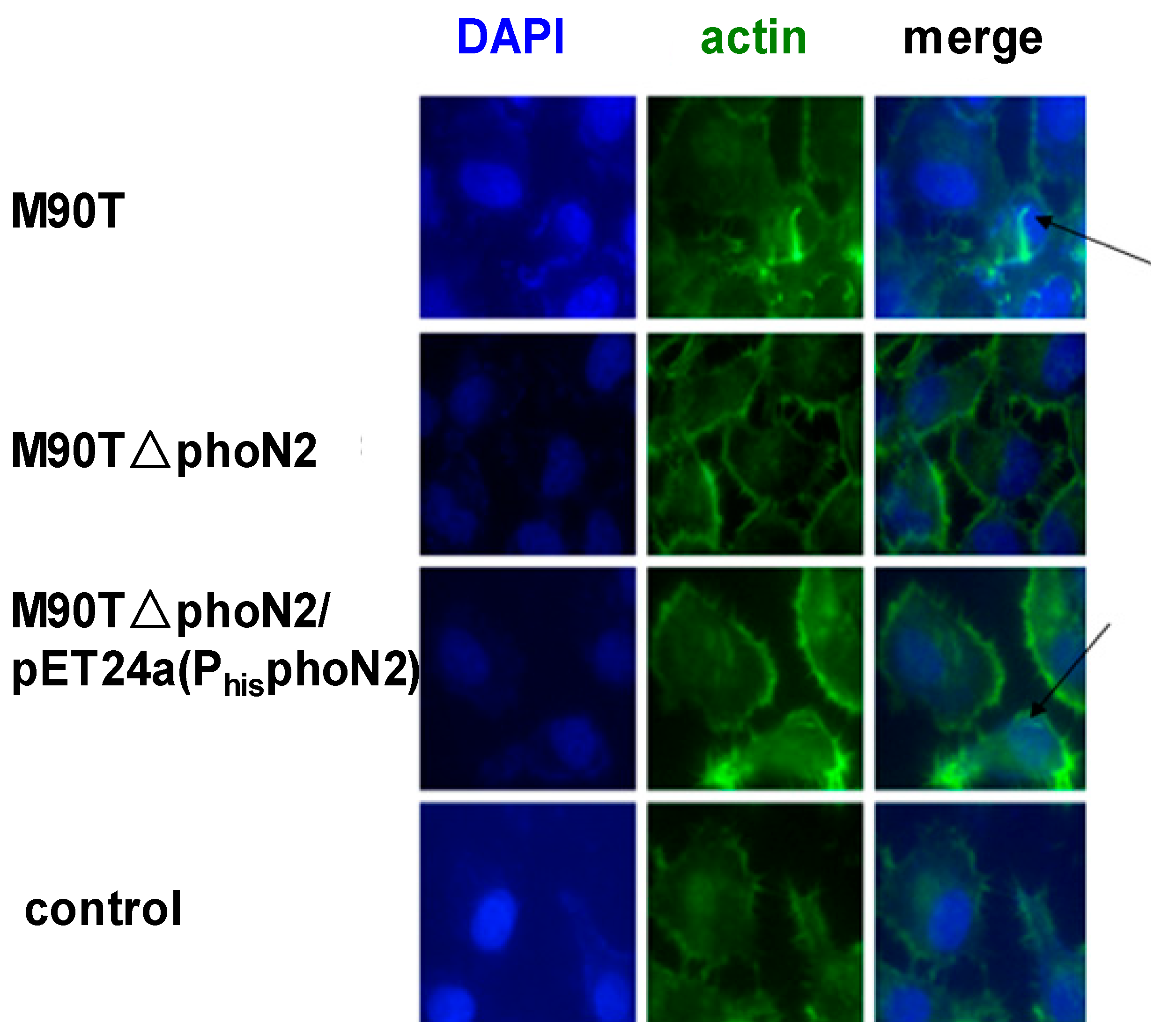
3. Experimental Section
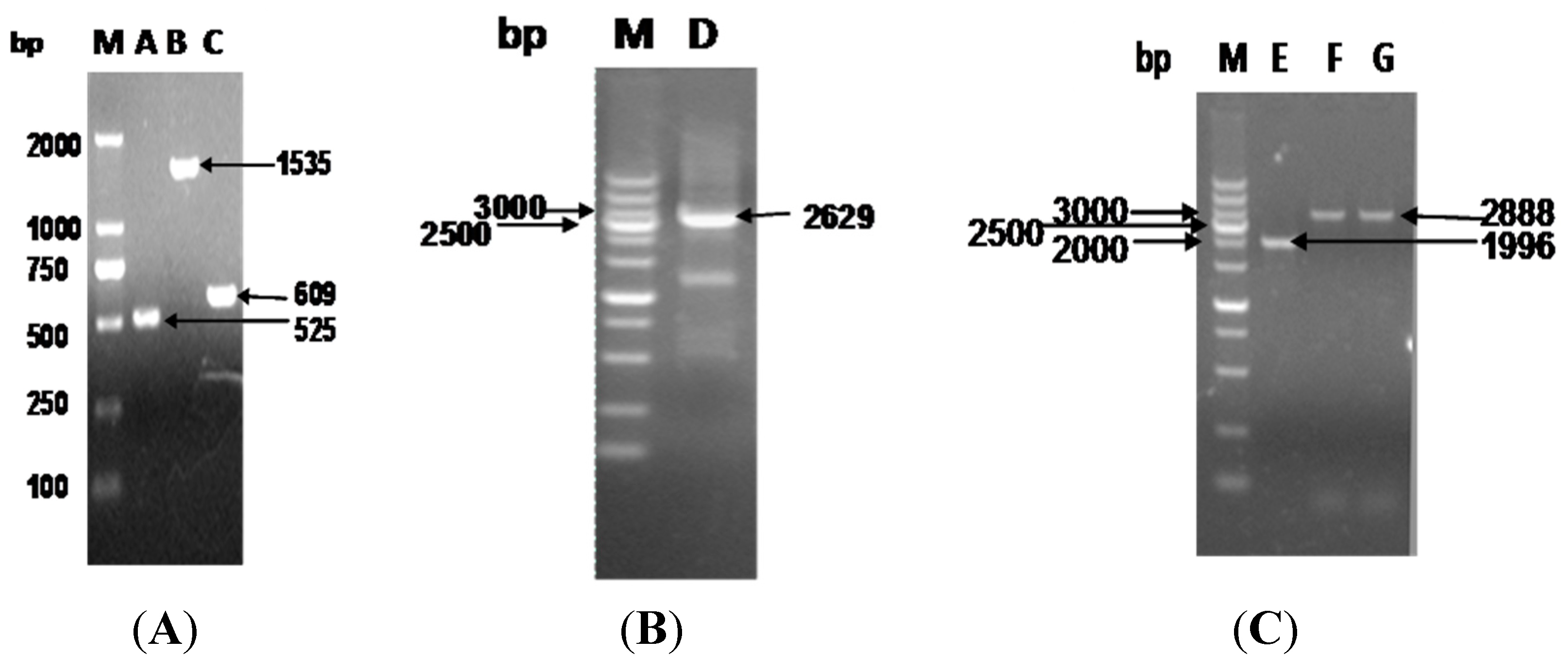
3.1. Construction of Recombinant Plasmids and Mutations in S. flexneri
| Name of Primers | Sequence of Primers |
|---|---|
| VirG1–1102 | F: 5'-GGATCCATGAATCAAATTCACAAATTTTTTT-3' |
| R: 5'-GCTCGAGTCAGAAGGTATATTTCACACCC-3' | |
| VirG53–779 | F: 5'-GGATCCATGACTCCTCTTTCGGGTA-3' |
| R: 5'-CTCGAGTCAGCGCCATGTGTGAATA-3' | |
| VirG758–1102 | F: 5'-CGGGATCCCGAGCTAGTTCACA-3' |
| R: 5'-CCGCTCGAGTCAGAAGGTATATTTCACA-3' | |
| VirG53–319 | F: 5'-GGATCCATGACTCCTCTTTCGGGTA-3' |
| R: 5'-CTCGAGTCATGTTCCATCATCTTCT-3' | |
| VirG320–507 | F: 5'-GGATCCATGCAAAATGTAGCAGGTA-3' |
| R: 5'-CCTCGAGTCAACTAACAGTAAGTTCA-3' | |
| VirG507–779 | F: 5'-GGATCCATGAGTACTATTCTGGCAG-3' |
| R: 5'-CTCGAGTCAGCGCCATGTGTGAATA-3' | |
| VirG591–758 | F: 5'-CGGGATCCAGTGGGACTGTGCTAAT-3' |
| R: 5'-CCGCTCGAGTCAGCGACTACTCATTTGA-3' | |
| phoN2 | F: 5'-AATTCCATATGATGAAAACCAAAAAC-3' |
| R: 5'-CTCGAGTGGGGTCAGTTCATTG-3' | |
| A-P1 | GGTCAAGAGCTTTTTCCCTACT |
| A-P2 | GCCTTCAGAGCATTTGCTGA |
| A-P3 | TAGCGAAGCCAAAAAAGAGT |
| A-P4 | ACTTAGTAAAGATGGTGCCAAC |
| A-P5 | TCAGCAAATGCTCTGAAGGCGTGTAGGCTGGAGCTGCTTC |
| A-P6 | ACTCTTTTTTGGCTTCGCTAATGGGAATTAGCCATGGTCC |
| A-PF | GCGTATCCTTAACTCTCTGCCTT |
| A-PR | GGCGATAAAAAAGCTGATATAGC |
3.2. Protein Expression
3.3. Purification of Proteins
3.4. Pull-Down Assays
3.5. Western Blot Analysis
3.6. Immunofluorescence Microscopy
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hale, T.L. Bacilliary dysentery. In Topley and Wilson’s Microbiology and Microbial Infections; John Wiley and Sons, Inc.: London, UK, 1998; Volume 3, pp. 479–493. [Google Scholar]
- Taylor, D.N.; Echeverria, P.; Sethabutr, O.; Pitarangsi, C.; Leksomboon, U.; Blacklow, N.R.; Rowe, B.; Gross, R.; Cross, J. Clinical and microbiologic features of Shigella and enteroinvasive Escherichia coli infections detected by DNA hybridization. J. Clin. Microbiol. 1988, 26, 1362–1366. [Google Scholar] [PubMed]
- Kothary, M.H.; Babu, U.S. Infective dose of foodborne pathogens in volunteers: A review. J. Food Saf. 2001, 21, 49–73. [Google Scholar] [CrossRef]
- Sankaran, K.; Banerjee, S.; Pavankumar, A.R.; Jesudason, M.; Reissbrodt, R.; Williams, P.H. Apyrase-based colorimetric test for detection of Shigella and enteroinvasive Escherichia coli in stool. Diagn. Microbiol. Infect. Dis. 2009, 63, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, G.N.; Hilbi, H. Molecular Pathogenesis of Shigella spp.: Controlling Host Cell Signaling, Invasion, and Death by Type III Secretion. Clin. Microbiol. Rev. 2008, 21, 134–156. [Google Scholar]
- Bernardini, M.L.; Mounier, J.; D’Hauteville, H.; Coquis-Rondon, M.; Sansonetti, P.J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 1989, 86, 3867–3871. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sasakawa, C. Molecular basis of the intracellular spreading of shigella. Infect. Immun. 2001, 69, 5959–5966. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.B.; Barzu, O.; Parsot, C.; Sansonetti, P.J. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J. Bacteriol. 1993, 175, 2189–2196. [Google Scholar] [PubMed]
- Sandlin, R.C.; Lampel, K.A.; Keasler, S.P.; Goldberg, M.B.; Stolzer, A.L.; Maurelli, A.T. Avirulence of rough mutants of Shigella flexneri: Requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect. Immun. 1995, 63, 229–237. [Google Scholar] [PubMed]
- Ansai, T.; Chen, X.; Barik, S.; Takehara, T. Conserved proline residues near the N-terminus are important for enzymatic activity of class A bacterial acid phosphatases. Arch. Biochem. Biophys. 2002, 408, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Sarli, S.; Nicoletti, M.; Schippa, S.; Del Chierico, F.; Santapaola, D.; Valenti, P.; Berlutti, F. Ala160 and His116 residues are involved in activity and specificity of apyrase, an ATP-hydrolysing enzyme produced by enteroinvasive Escherichia coli. Microbiology 2005, 151, 2853–2860. [Google Scholar] [CrossRef] [PubMed]
- Santapaola, D.; Del Chierico, F.; Petrucca, A.; Uzzau, S.; Casalino, M.; Colonna, B.; Sessa, R.; Berlutti, F.; Nicoletti, M. Apyrase, the Product of the Virulence Plasmid-Encoded phoN2 (apy)Gene of Shigella flexneri, Is Necessary for Proper Unipolar IcsA Localization and for Efficient Intercellular Spread. J. Bacteriol. 2006, 188, 1620–1627. [Google Scholar] [CrossRef] [PubMed]
- Scribano, D.; Petrucca, A.; Pompili, M.; Ambrosi, C.; Bruni, E.; Zagaglia, C.; Prosseda, G.; Nencioni, L.; Casalino, M.; Polticelli, F.; et al. Polar localization of PhoN2, a periplasmic virulence-associated factor of Shigella flexneri, is required for proper IcsA exposition at the old bacterial pole. PLoS One 2014, 9, e90230. [Google Scholar]
- Varjosalo, M.; Sacco, R.; Stukalov, A.; Drogen, A.V.; Planyavsky, M.; Hauri, S.; Aebersold, R.; Bennett, K.L.; Colinge, J.; Gstaiger, M.; et al. Inerlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods 2013, 10, 307–314. [Google Scholar]
- Huang, J.; Niu, C.; Green, C.D.; Yang, L.; Mei, H.; Han, J.D. Systematic prediction of pharmacodynamics drug-drug interactions through protein-protein-interaction network. PLoS Comput. Biol. 2013, 9, e1002998. [Google Scholar]
- Yasui, N.; Findlay, G.M.; Gish, G.D.; Hsiung, M.S.; Huang, J.; Tucholska, M.; Taylor, L.; Smith, L.; Boldridge, W.C.; Koide, A.; et al. Directed network wiring identifies a key protein interaction in embryonic stem cell differentiation. Mol. Cell 2014, 54, 1034–1041. [Google Scholar]
- Steiner, S.; Lori, C.; Boehm, A.; Jenal, U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2012, 32, 354–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, S.; Solano, R. Pull-down analysis of interactions among jasmonic acid core signaling proteins. Methods Mol. Biol. 2013, 1011, 159–171. [Google Scholar] [PubMed]
- Mochizuki, Y.; Kohno, F.; Nishigaki, K.; Nemoto, N. A pull-down method with a biotinylated bait protein prepared by cell-free translation using a puromycin linker. Anal. Biochem. 2013, 434, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Abascal-Palacios, G.; Schindler, C.; Rojas, A.L.; Bonifacino, J.S.; Hierro, A. Structural basis for the interaction of the golgi-associated retrograde protein complex with the t-SNARE syntaxin 6. Structure 2013, 21, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Klauser, T.; Pohlner, J.; Meyer, T.F. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease β-domain: Conformation-dependent outer membrane translocation. EMBO J. 1990, 9, 1991–1999. [Google Scholar] [PubMed]
- Suzuki, T.; Miki, H.; Takenawa, T.; Sasakawa, C. Neural Wiskott-Aldrich syndrome protein is implicated in actin-based motility of Shigella flexneri. EMBO J. 1998, 17, 2767–2776. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Saga, S.; Sasakawa, C. Functional analysis of Shigella VirG domains essential for interaction with vinculin and actin-based motility. J. Biol. Chem. 1996, 271, 21878–21885. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Janakiraman, A.; Fixen, K.R.; Gray, A.N. A genome-scale proteomic screen identifies a role for DnaK in chaperoning of polar autotranspoters in Shigella. J. Bacteriol. 2009, 191, 6300–6311. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Gong, G.-H.; Zhou, W.; Zhang, B.; Bao, S.-Y.; Wei, C.-X.; Yue, J.-J.; Zhang, Y.-F. Analysis on the Interaction Domain of VirG and Apyrase by Pull-Down Assay. Molecules 2014, 19, 18090-18101. https://doi.org/10.3390/molecules191118090
Wang Y, Gong G-H, Zhou W, Zhang B, Bao S-Y, Wei C-X, Yue J-J, Zhang Y-F. Analysis on the Interaction Domain of VirG and Apyrase by Pull-Down Assay. Molecules. 2014; 19(11):18090-18101. https://doi.org/10.3390/molecules191118090
Chicago/Turabian StyleWang, Yu, Guo-Hua Gong, Wei Zhou, Bin Zhang, Shu-Yin Bao, Cheng-Xi Wei, Jun-Jie Yue, and Yan-Fen Zhang. 2014. "Analysis on the Interaction Domain of VirG and Apyrase by Pull-Down Assay" Molecules 19, no. 11: 18090-18101. https://doi.org/10.3390/molecules191118090





