Structural Probes in Quadruplex Nucleic Acid Structure Determination by NMR
Abstract
:1. Introduction
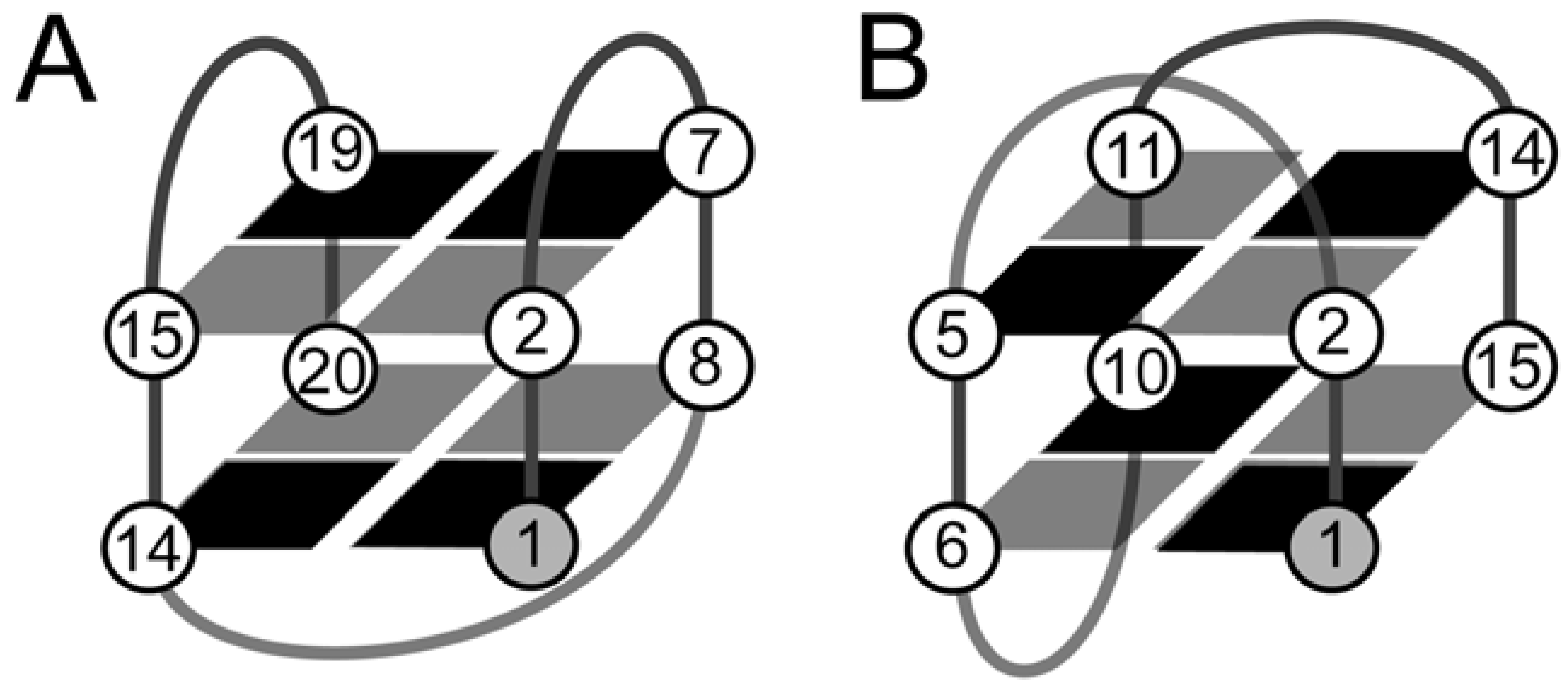
2. The Structure of Quadruplexes
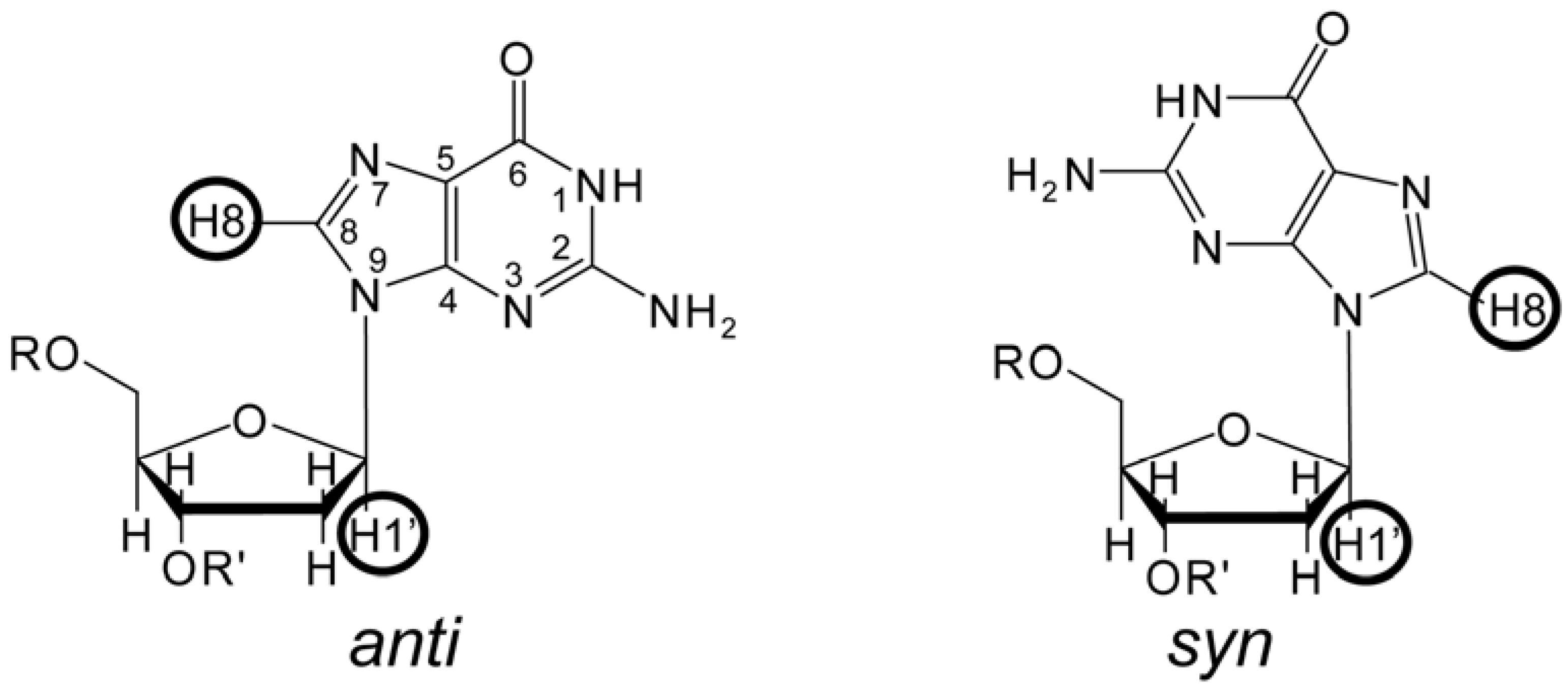
3. The Identification of NMR Signals
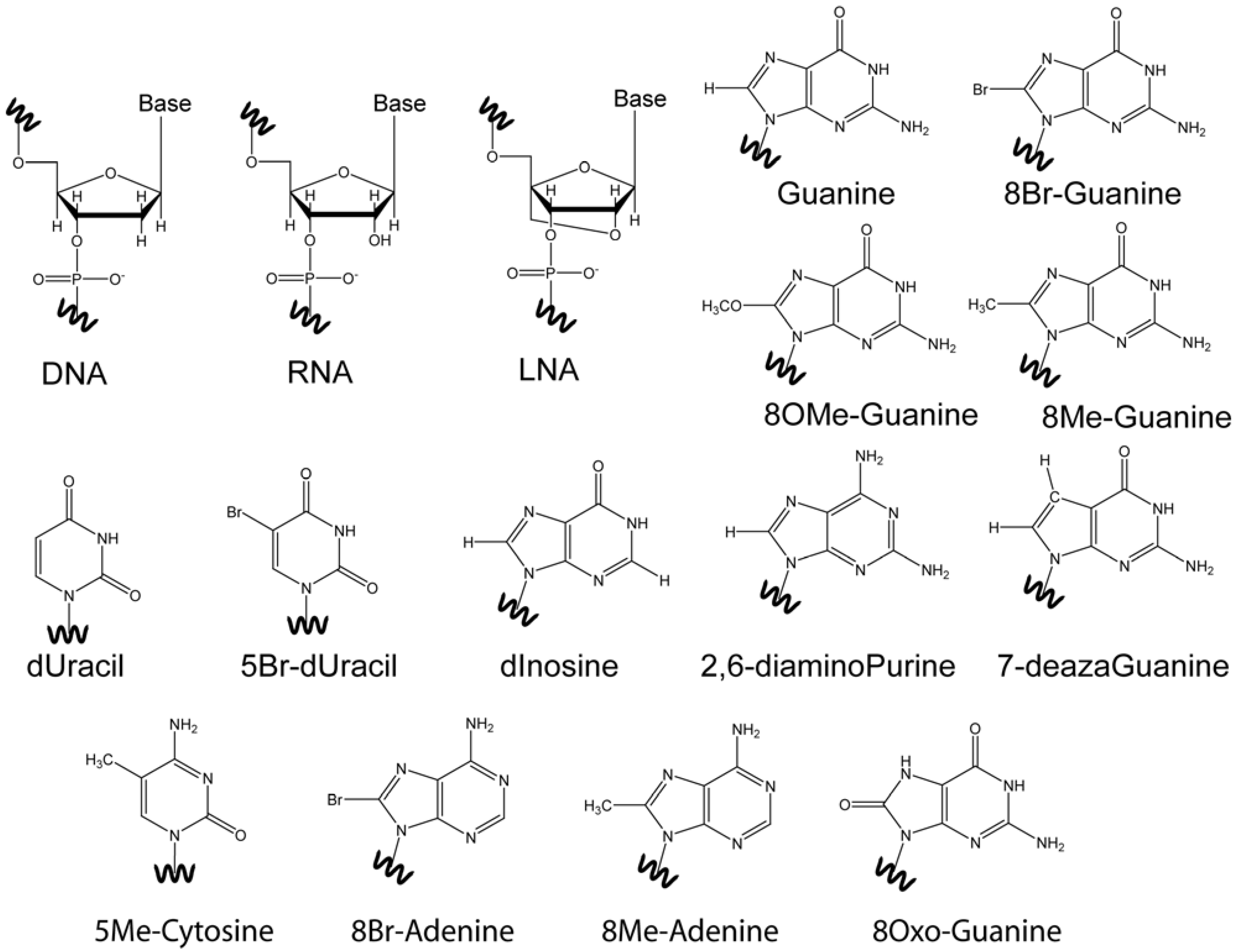
4. Base Modifications
4.1. 8-Br-Guanosine Substitutions Prevent Formation of Narrow Grooves
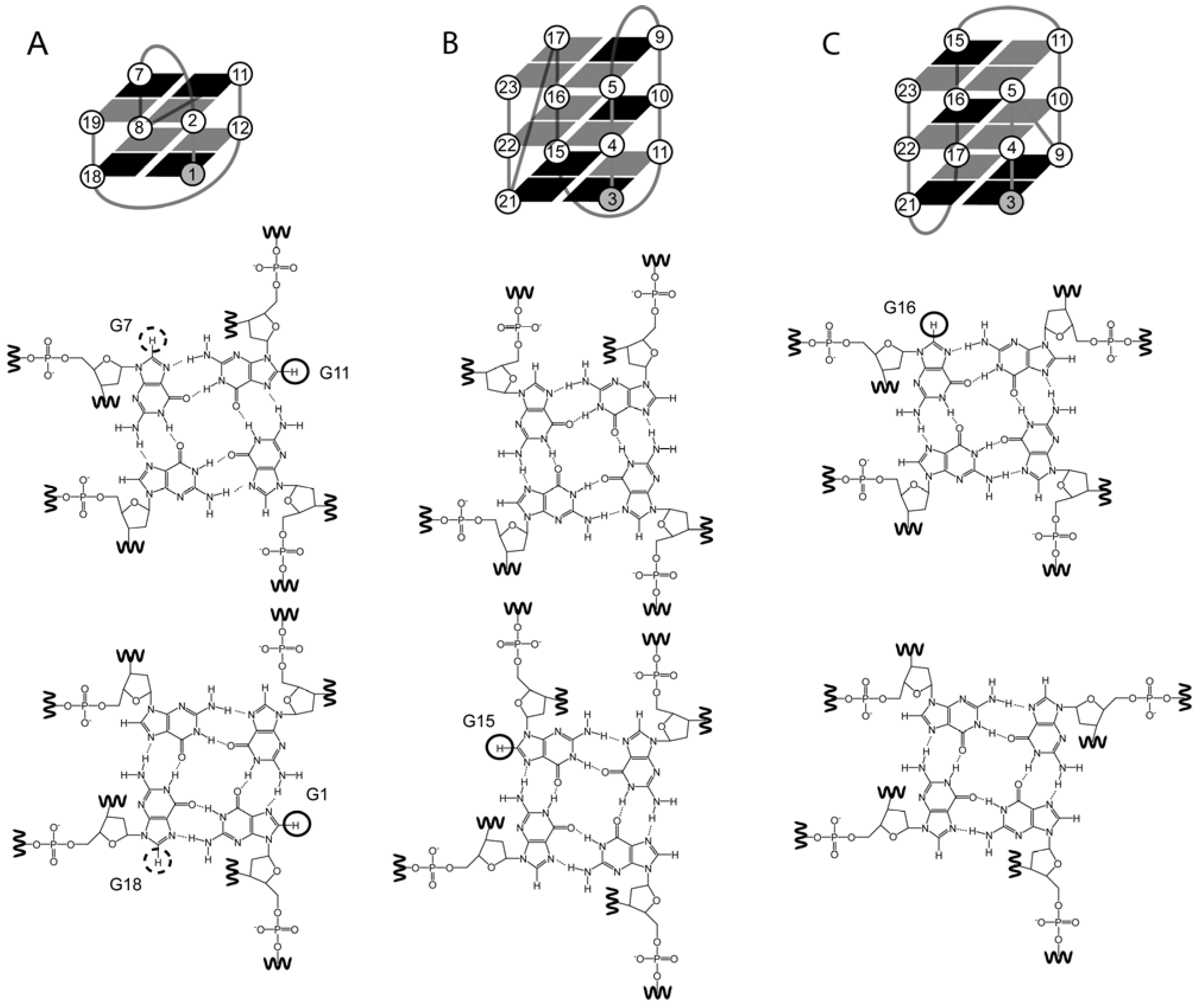
4.2. The Effect of O-Methyl, Amino, and Methyl C8 Substitutions
4.3. The Effect of 8Oxo-Guanine C8 Substitutions
4.4. Base Modifications in Thymine, Cytosine, and Adenine
4.5. Abasic Sites
5. Ribo-Base Modifications in DNA and Vice-Versa
6. Backbone Modifications
7. Inosine Studies
8. Isotopic Enrichment
9. Perspectives
Acknowledgments
References
- Webba da Silva, M. NMR methods for studying quadruplex nucleic acids. Methods 2007, 43, 264–277. [Google Scholar] [CrossRef]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR spectroscopy of G-quadruplexes. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef]
- Musumeci, D.; Montesarchio, D. Polyvalent nucleic acid aptamers and modulation of their activity: A focus on the thrombin binding aptamer. Pharmacol. Ther. 2012, 136, 202–215. [Google Scholar] [CrossRef]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin Binding Aptamer, More than a Simple Aptamer: Chemically Modified Derivatives and Biomedical Applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef]
- Webba da Silva, M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007, 13, 9738–9745. [Google Scholar] [CrossRef]
- Lech, C.J.; Lim, J.K.C.; Lim, J.M.W.; Amrane, S.; Heddi, B.; Phan, A.T. Effects of Site-Specific Guanine C8-Modifications on an Intramolecular DNA G-Quadruplex. Biophys. J. 2011, 101, 1987–1998. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Majumdar, A.; Shallop, A.; Chernichenko, N.; Skripkin, E.; Jones, R.; Patel, D.J. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)center dot G(syn)center dot G(anti)center dot G(anti) tetrads flanked by a G center dot(T-T) triad and a T center dot T center dot T triple. J. Mol. Biol. 2001, 310, 181–194. [Google Scholar]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar]
- Matsugami, A.; Xu, Y.; Noguchi, Y.; Sugiyama, H.; Katahira, M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007, 274, 3545–3556. [Google Scholar]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the Human Telomere in K+ Solution: A Stable Basket-Type G-Quadruplex with Only Two G-Tetrad Layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar]
- Sugiyama, H.; Kawai, K.; Matsunaga, A.; Fujimoto, K.; Saito, I.; Robinson, H.; Wang, A.H.J. Synthesis, structure and thermodynamic properties of 8-methylguanine-containing oligonucleotides: Z-DNA under physiological salt conditions. Nucleic Acids Res. 1996, 24, 1272–1278. [Google Scholar]
- Krawczyk, S.H.; Bischofberger, N.; Griffin, L.C.; Law, V.S.; Shea, R.G.; Swaminathan, S. Structure-activity study of oligodeoxynucleotides which inhibit thrombin. Nucleosides Nucleotides 1995, 14, 1109–1116. [Google Scholar] [CrossRef]
- He, G.X.; Krawczyk, S.H.; Swaminathan, S.; Shea, R.G.; Dougherty, J.P.; Terhorst, T.; Law, V.S.; Griffin, L.C.; Coutre, S.; Bischofberger, N. N-2- and C-8-substituted oligodeoxynucleotides with enhanced thrombin inhibitory activity in vitro and in vivo. J. Med. Chem. 1998, 41, 2234–2242. [Google Scholar]
- Virgilio, A.; Esposito, V.; Citarella, G.; Pepe, A.; Mayol, L.; Galeone, A. The insertion of two 8-methyl-2'-deoxyguanosine residues in tetramolecular quadruplex structures: Trying to orientate the strands. Nucleic Acids Res. 2012, 40, 461–475. [Google Scholar]
- Phong, L.T.T.; Virgilio, A.; Esposito, V.; Citarella, G.; Mergny, J.L.; Galeone, A. Effects of 8-methylguanine on structure, stability and kinetics of formation of tetramolecular quadruplexes. Biochimie 2011, 93, 399–408. [Google Scholar]
- Cosconati, S.; Marinelli, L.; Trotta, R.; Virno, A.; de Tito, S.; Romagnoli, R.; Pagano, B.; Limongelli, V.; Giancola, C.; Baraldi, P.G.; et al. Structural and Conformational Requisites in DNA Quadruplex Groove Binding: Another Piece to the Puzzle. J. Am. Chem. Soc. 2010, 132, 6425–6433. [Google Scholar]
- Martino, L.; Virno, A.; Pagano, B.; Virgilio, A.; Di Micco, S.; Galeone, A.; Giancola, C.; Bifulco, G.; Mayol, L.; Randazzo, A. Structural and thermodynamic studies of the interaction of distamycin a with the parallel quadruplex structure d(TGGGGT) (4). J. Am. Chem. Soc. 2007, 129, 16048–16056. [Google Scholar]
- Xu, Y.; Sugiyama, H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 2006, 34, 949–954. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Kettani, A.; Wang, W.M.; Jones, R.; Patel, D.J. A diamond-shaped zipper-like DNA architecture containing triads sandwiched between mismatches and tetrads. J. Mol. Biol. 2000, 295, 455–469. [Google Scholar] [CrossRef]
- Virgilio, A.; Esposito, V.; Randazzo, A.; Mayol, L.; Galeone, A. Effects of 8-methyl-2'-deoxyadenosine incorporation into quadruplex forming oligodeoxyribonucleotides. Bioorg. Med. Chem. 2005, 13, 1037–1044. [Google Scholar] [CrossRef]
- Phan, A.T.; Patel, D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef]
- Amrane, S.; Ang, R.W.L.; Tan, Z.M.; Li, C.; Lim, J.K.C.; Lim, J.M.W.; Lim, K.W.; Phan, A.T. A novel chair-type G-quadruplex formed by a Bombyx mori telomeric sequence. Nucleic Acids Res. 2009, 37, 931–938. [Google Scholar] [CrossRef]
- Cevec, M.; Plavec, J. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry 2005, 44, 15238–15246. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Brown, T.; Fox, K.R. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007, 581, 1657–1660. [Google Scholar]
- Esposito, V.; Martino, L.; Citarella, G.; Virgilio, A.; Mayol, L.; Giancola, C.; Galeone, A. Effects of abasic sites on structural, thermodynamic and kinetic properties of quadruplex structures. Nucleic Acids Res. 2010, 38, 2069–2080. [Google Scholar]
- Skolakova, P.; Bednarova, K.; Vorlickova, M.; Sagi, J. Quadruplexes of human telomere dG(3)(TTAG(3))(3) sequences containing guanine abasic sites. Biochem. Biophys. Res. Commun. 2010, 399, 203–208. [Google Scholar] [CrossRef]
- Virgilio, A.; Petraccone, L.; Esposito, V.; Citarella, G.; Giancola, C.; Galeone, A. The abasic site lesions in the human telomeric sequence d[TA(G(3)T(2)A)(3)G(3)]: A thermodynamic point of view. Biochim. Biophys. Acta 2012, 1820, 2037–2043. [Google Scholar]
- Tang, C.F.; Shafer, R.H. Engineering the quadruplex fold: Nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006, 128, 5966–5973. [Google Scholar] [CrossRef]
- Martadinata, H.; Phan, A.T. Structure of Propeller-Type Parallel-Stranded RNA G-Quadruplexes, Formed by Human Telomeric RNA Sequences in K+ Solution. J. Am. Chem. Soc. 2009, 131, 2570–2578. [Google Scholar] [CrossRef]
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630. [Google Scholar] [CrossRef]
- Singh, S.K.; Nielsen, P.; Koshkin, A.A.; Wengel, J. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 455–456. [Google Scholar]
- Pradhan, D.; Hansen, L.H.; Vester, B.; Petersen, M. Selection of G-Quadruplex Folding Topology with LNA-Modified Human Telomeric Sequences in K+ Solution. Chem. Eur. J. 2011, 17, 2405–2413. [Google Scholar]
- Braasch, D.A.; Corey, D.R. Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001, 8, 1–7. [Google Scholar] [CrossRef]
- Dominick, P.K.; Jarstfer, M.B. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004, 126, 5050–5051. [Google Scholar]
- Randazzo, A.; Esposito, V.; Ohlenschlager, O.; Ramachandran, R.; Mayol, L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004, 32, 3083–3092. [Google Scholar] [CrossRef]
- Nielsen, J.T.; Arar, K.; Petersen, M. NMR solution structures of LNA (locked nucleic acid) modified quadruplexes. Nucleic Acids Res. 2006, 34, 2006–2014. [Google Scholar]
- Virno, A.; Randazzo, A.; Giancola, C.; Bucci, M.; Cirinoc, G.; Mayol, L. A novel thrombin binding aptamer containing a G-LNA residue. Bioorg. Med. Chem. 2007, 15, 5710–5718. [Google Scholar] [CrossRef]
- Nielsen, J.T.; Arar, K.; Petersen, M. Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology. Angew. Chem. Int. Ed. Engl. 2009, 48, 3099–3103. [Google Scholar] [CrossRef]
- Smith, F.W.; Feigon, J. Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry 1993, 32, 8682–8692. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the tetrahymena telomeric repeat d(T(2)G(4))(4) G-tetraplex. Structure 1994, 2, 1141–1156. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Ngoc Quang, D.; Phan, A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011, 39, 8984–8991. [Google Scholar] [CrossRef]
- Hu, L.; Lim, K.W.; Bouaziz, S.; Phan, A.T. Giardia Telomeric Sequence d(TAGGG)4 Forms Two Intramolecular G-Quadruplexes in K+ Solution: Effect of Loop Length and Sequence on the Folding Topology. J. Am. Chem. Soc. 2009, 131, 16824–16831. [Google Scholar] [CrossRef]
- Lim, K.W.; Lacroix, L.; Yue, D.J.E.; Lim, J.K.C.; Lim, J.M.W.; Phan, A.T. Coexistence of Two Distinct G-Quadruplex Conformations in the hTERT Promoter. J. Am. Chem. Soc. 2010, 132, 12331–12342. [Google Scholar] [CrossRef]
- Yue, D.J.E.; Lim, K.W.; Phan, A.T. Formation of (3+1) G-Quadruplexes with a Long Loop by Human Telomeric DNA Spanning Five or More Repeats. J. Am. Chem. Soc. 2011, 133, 11462–11465. [Google Scholar] [CrossRef]
- Zhang, N.; Gorin, A.; Majumdar, A.; Kettani, A.; Chernichenko, N.; Skripkin, E.; Patel, D.J. V-shaped scaffold: A new architectural motif identified in an A center dot(G center dot G center dot G center dot G) pentad-containing dimeric DNA quadruplex involving stacked G(anti)center dot G(anti)center dot G(anti)center dot G(syn) tetrads. J. Mol. Biol. 2001, 311, 1063–1079. [Google Scholar] [CrossRef]
- Zhang, N.; Phan, A.T.; Patel, D.J. (3+1) assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005, 127, 17277–17285. [Google Scholar] [CrossRef]
- Dai, J.X.; Chen, D.; Jones, R.A.; Hurley, L.H.; Yang, D.Z. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006, 34, 5133–5144. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.X.; Jones, R.A.; Yang, D.Z. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar]
- Ambrus, A.; Chen, D.; Dai, J.X.; Bialis, T.; Jones, R.A.; Yang, D.Z. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Kettani, A.; Bouaziz, S.; Wang, W.; Jones, R.A.; Patel, D.J. Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads. Nat. Struct. Biol. 1997, 4, 382–389. [Google Scholar] [CrossRef]
- Kettani, A.; Bouaziz, S.; Gorin, A.; Zhao, H.; Jones, R.A.; Patel, D.J. Solution structure of a Na cation stabilized DNA quadruplex containing G center dot G center dot G center dot G and G center dot C center dot G center dot C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998, 282, 619–636. [Google Scholar] [CrossRef]
- Gaffney, B.L.; Wang, C.; Jones, R.A. Nitrogen-15-labeled oligodeoxynucleotides. 4. Tetraplex formation of d[G(15N7)GTTTTTGG] and d[T(15N7)GGGT] monitored by 1H detected 15N NMR. J. Am. Chem. Soc. 1992, 114, 4047–4050. [Google Scholar] [CrossRef]
- Gaffney, B.L.; Kung, P.P.; Wang, C.; Jones, R.A. Nitrogen-15-Labeled Oligodeoxynucleotides. 8. Use of 15N NMR To Probe Hoogsteen Hydrogen-Bonding at Guanine and Adenine N7 Atoms of a DNA Triplex. J. Am. Chem. Soc. 1995, 117, 12281–12283. [Google Scholar] [CrossRef]
- Otting, G.; Wüthrich, K. Heteronuclear filters in two-dimensional [1H, 1H]-NMR-spectroscopy: Combined use with isotope labeling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 1990, 23, 39–96. [Google Scholar] [CrossRef]
- Zhang, N.; Gorin, A.; Majumdar, A.; Kettani, A.; Chernichenko, N.; Skripkin, E.; Patel, D.J. Dimeric DNA quadruplex containing major groove-aligned A center dot T center dot A center dot T and G center dot C center dot G center dot C tetrads stabilized by inter-subunit Watson-Crick A center dot T and G center dot C pairs. J. Mol. Biol. 2001, 312, 1073–1088. [Google Scholar] [CrossRef]
- Sotoya, H.; Matsugami, A.; Ikeda, T.; Ouhashi, K.; Uesugi, S.; Katahira, M. Method for direct discrimination of intra- and intermolecular hydrogen bonds, and characterization of the G(:A):G(:A):G(:A):G heptad, with scalar couplings across hydrogen bonds. Nucleic Acids Res. 2004, 32, 5113–5118. [Google Scholar]
- Phan, A.T.; Patel, D.J. A site-specific low-enrichment N-15,C-13 isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002, 124, 1160–1161. [Google Scholar] [CrossRef]
- Phan, A.T.; Patel, D.J. Differentiation between unlabeled and very-low-level fully N-15,C-13-labeled nucleotides for resonance assignments in nucleic acids. J. Biomol. NMR 2002, 23, 257–262. [Google Scholar] [CrossRef]
- Phan, A.T.; Modi, Y.S.; Patel, D.J. Propeller-type parallel-stranded g-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004, 126, 8710–8716. [Google Scholar] [CrossRef]
- Phan, A.T.; Modi, Y.S.; Patel, D.J. Two-repeat Tetrahymena telomeric d(TGGGGTTGGGGT) sequence interconverts between asymmetric dimeric G-quadruplexes in solution. J. Mol. Biol. 2004, 338, 93–102. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Karsisiotis, A.I.; Webba da Silva, M. Structural Probes in Quadruplex Nucleic Acid Structure Determination by NMR. Molecules 2012, 17, 13073-13086. https://doi.org/10.3390/molecules171113073
Karsisiotis AI, Webba da Silva M. Structural Probes in Quadruplex Nucleic Acid Structure Determination by NMR. Molecules. 2012; 17(11):13073-13086. https://doi.org/10.3390/molecules171113073
Chicago/Turabian StyleKarsisiotis, Andreas Ioannis, and Mateus Webba da Silva. 2012. "Structural Probes in Quadruplex Nucleic Acid Structure Determination by NMR" Molecules 17, no. 11: 13073-13086. https://doi.org/10.3390/molecules171113073
APA StyleKarsisiotis, A. I., & Webba da Silva, M. (2012). Structural Probes in Quadruplex Nucleic Acid Structure Determination by NMR. Molecules, 17(11), 13073-13086. https://doi.org/10.3390/molecules171113073




