DNA-Directed Base Pair Opening
Abstract
:1. Introduction
2. Results and Discussion
2.1. DNA Crossovers

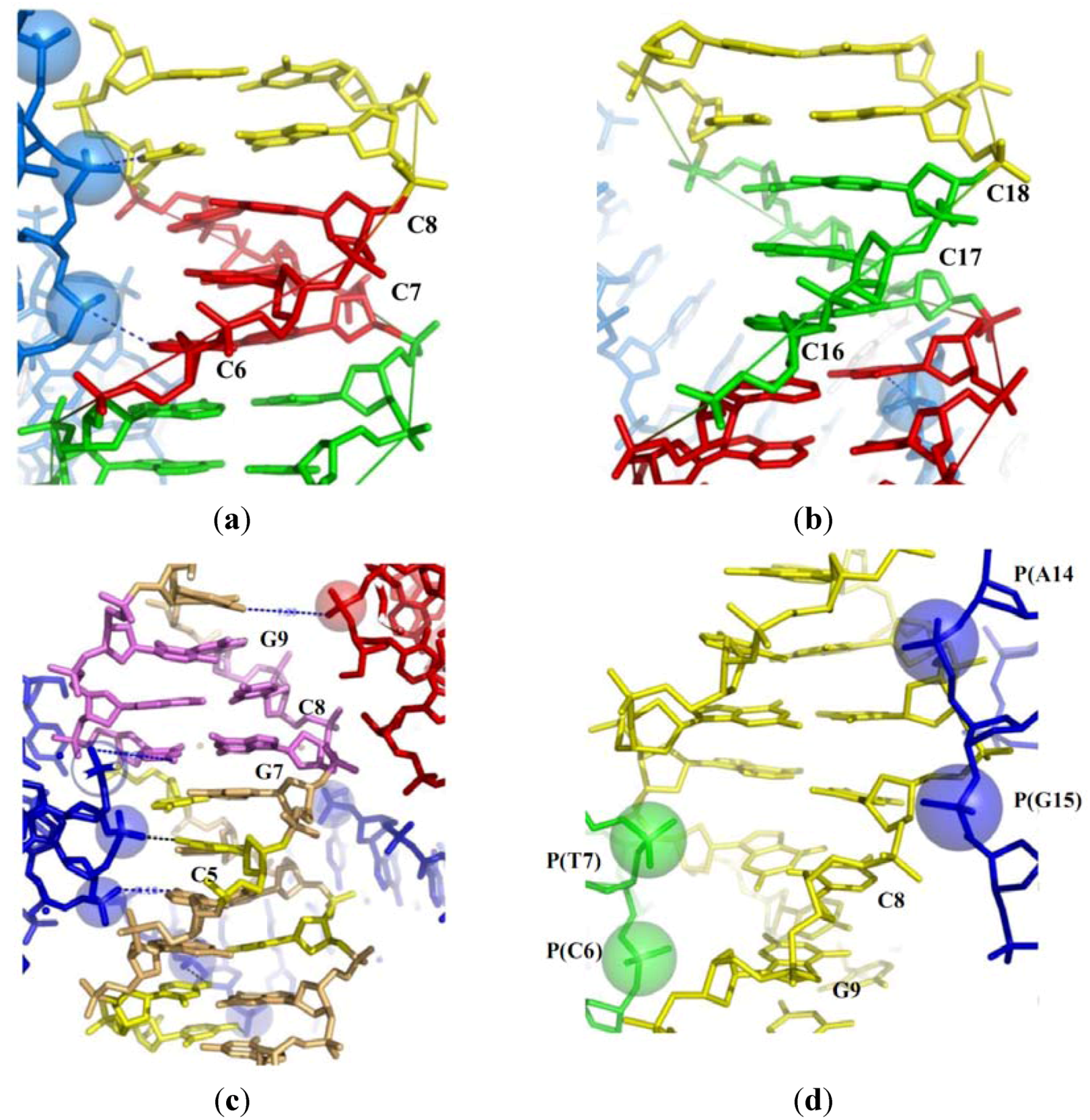
2.2. Tight Contacts in DNA Higher-Order Structures and Supercoiling
2.3. DNA-Directed Base-Pair Opening
2.3.1. Close Helical Approach and Base Stacking Alterations
| Bdjb77 | Bd0015 | Bd0084 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prop. | Buckle | Dist. | Prop. | Buckle | Dist. | Prop | Buckle | Dist. | |||
| (°) | (°) | P-axis | (°) | (°) | P-axis | (°) | (°) | P-axis | |||
| (Å) | (Å) | (Å) | |||||||||
| C1-G20 | −5.2 | 10.2 | C1-G20 | −8.7 | 6.8 | P(15) | C1-G20 | −11.6 | −0.4 | ||
| 8.2 | |||||||||||
| C2-G19 | 0.6 | 1.4 | C2-G19 | −9.9 | −6.6 | P(14) | C2-G19 | 15.6 | 1.4 | ||
| 6.6 | |||||||||||
| I3-C18 | −10.1 | −0.5 | G3-C18 | −14.5 | −12.2 | G3-C18 | −7.0 | 1.4 | |||
| I4-C17 | −6.8 | −2.6 | C4-G17 | −30.6 | 1.8 | P(18) | A4-T17 | 5.7 | −3.3 | ||
| 8.1 | |||||||||||
| I5-C16 | −12.4 | 1.9 | C5-G16 | − 9.2 | 6.5 | P(17) | G5-C16 | −24.5 | −5.4 | P(A14) | |
| 6.9 | 9.8 | ||||||||||
| C6-I15 | −15.2 | −0.7 | P(10) | G6-C15 | −11.4 | −1.8 | C6-G15 | −11.0 | 1.2 | ||
| 7.5 | |||||||||||
| C7-I14 | −15.3 | 11.6 | G7-C14 | −26.1 | 0.3 | T7-A14 | −5.8 | 2.0 | |||
| C8-I13 | −13.6 | 16.8 | C8-G13 | −14.0 | −3.5 | C8-G13 | −12 | 2.0 | P(T7) | ||
| 6.8 | |||||||||||
| G9-C12 | −6.1 | 4.8 | P(9) | G9-C12 | −28.7 | 3.3 | G9-C12 | −23.3 | −23.4 | P(C6) | |
| 8.1 | 8.0 | ||||||||||
| G10-C11 | −17.3 | 1.6 | G10-C11 | −16.8 | −4.7 | G10-C11 | −14.9 | 6.3 | |||
2.3.2. Close Helical Approach and Base Pair Opening
2.3.2.1. Direct Base-Pair Opening
| Space group | Resol. | Sequence | Structural alteration | |
|---|---|---|---|---|
| Base unstacking | ||||
| Bdjb77 (286D) | P31 | 2.5 | 5'-CC I I I CCCGG | High propeller twist and buckle values (from C6-I15 to C8 I13) in front of inserted phosphate groups (Table 1) |
| 3'-GGCCC I I I CC | ||||
| Bd0015 (1QC1) | H 3 | 2.5 | 5'-CCGCCGGCGG | Alterations of base stacking along the whole structure (Table 1) |
| 3'-GGCGGCCGCC | ||||
| Bd0084 (1ZFG) | P31 | 1.75 | 5'-CCGAGCTCGG | Alterations of base stacking in front of the inserted phosphate groups (Table 1) |
| 3'-GGCTCGAGCC | ||||
| Direct base-pair opening | ||||
| Bd0022 (1QP5) | H 3 | 2.6 | * * | C3-G17(#): N4-O1P: 2.4 Å |
| C21-G17(#): N4-O1P: 3.3 Å | ||||
 | C18-C20(#): N4-O1P: 2.9 Å | |||
 | ||||
| * * | Shift of the base pairing in the major groove (Figure 5) from  | |||
| Bdl035 (330D) | H 3 | 2.7 | 5'-ACCGCCGGCGCC | Normal base-pairing |
| 3'-TGGCGGCCGCGG | ||||
| Bd0076 (1ZEW) | C2 | 2.25 |  | C12-G7(#): N4-O1P: 2.7 Å |
 : N4-O6: 4.0 Å : N4-O6: 4.0 Å | ||||
| N3-N1: 3.3 Å | ||||
| O2-N2: 2.7 Å | ||||
 |  : O4-N6: 3.6 Å : O4-N6: 3.6 Å | |||
| * | N3-N1: 3.6 Å | |||
| A18-T15(#) N6-O1P: 6.9Å | ||||
 : N6-O4 : 4.0 Å : N6-O4 : 4.0 Å | ||||
| N1-N3: 3.7 Å | ||||
| Bd0028 (1ZFH) | C2 | 2.5 | 5'-CCGCTAGCGG | Normal base-pairing |
| 3'-GGCGATCGCC | ||||
| Bd0021 (1CW9) | P212121 | 1.55 | * | C6-G10(#): N4-O1P: 3 Å |
 |  : N4-O6: 7.0 Å : N4-O6: 7.0 Å | |||
| 3'-GGTCCGGACC | (2 mol/au: open and closed form) | |||
| Indirect base-pair opening | ||||
| Bdf062 (206D) | P6122 | 2.5 | * | C1-C8(#): N4-O1P: 3.3 Å |
| C10-A9(#): C6-O1P: 2.9 Å | ||||
 | C5-O2P: 3.3 Å | |||
 : O4-N6: 4.7 Å : O4-N6: 4.7 Å | ||||
 | O2-N6: 2.9 Å | |||
| Unstacking C1/G2/G3 | ||||
| Ud0029 (1P4Z) | P6122 | 2 | * | C7-A6(#): N4-O1P: 3.0 Å |
 | N4-O6: 3.3 Å | |||
 |  N6-O4: 3.8 Å N6-O4: 3.8 Å | |||
| * | N1-N3: 3.5 Å | |||
| Bd0068 (1S1K) | P6122 | 1.9 | 5'-CCMGTACTGG | Normal base-pairing |
| 3'-GGTCATGMCC | ||||
| M=1AP | ||||



2.3.2.2. Indirect Base-Pair Opening
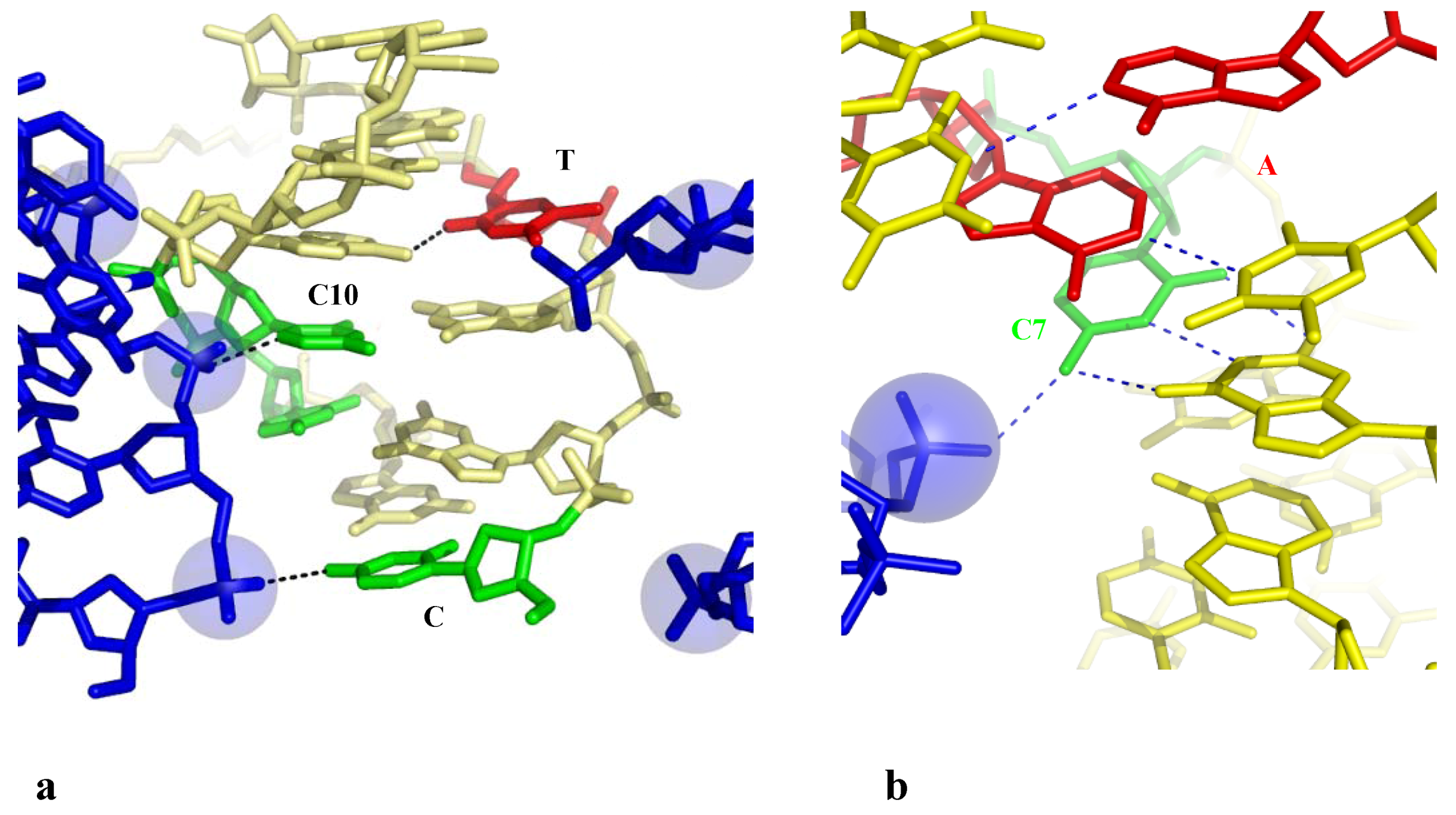
2.4. A Model for the Initiation of DNA Melting Induced by Compaction
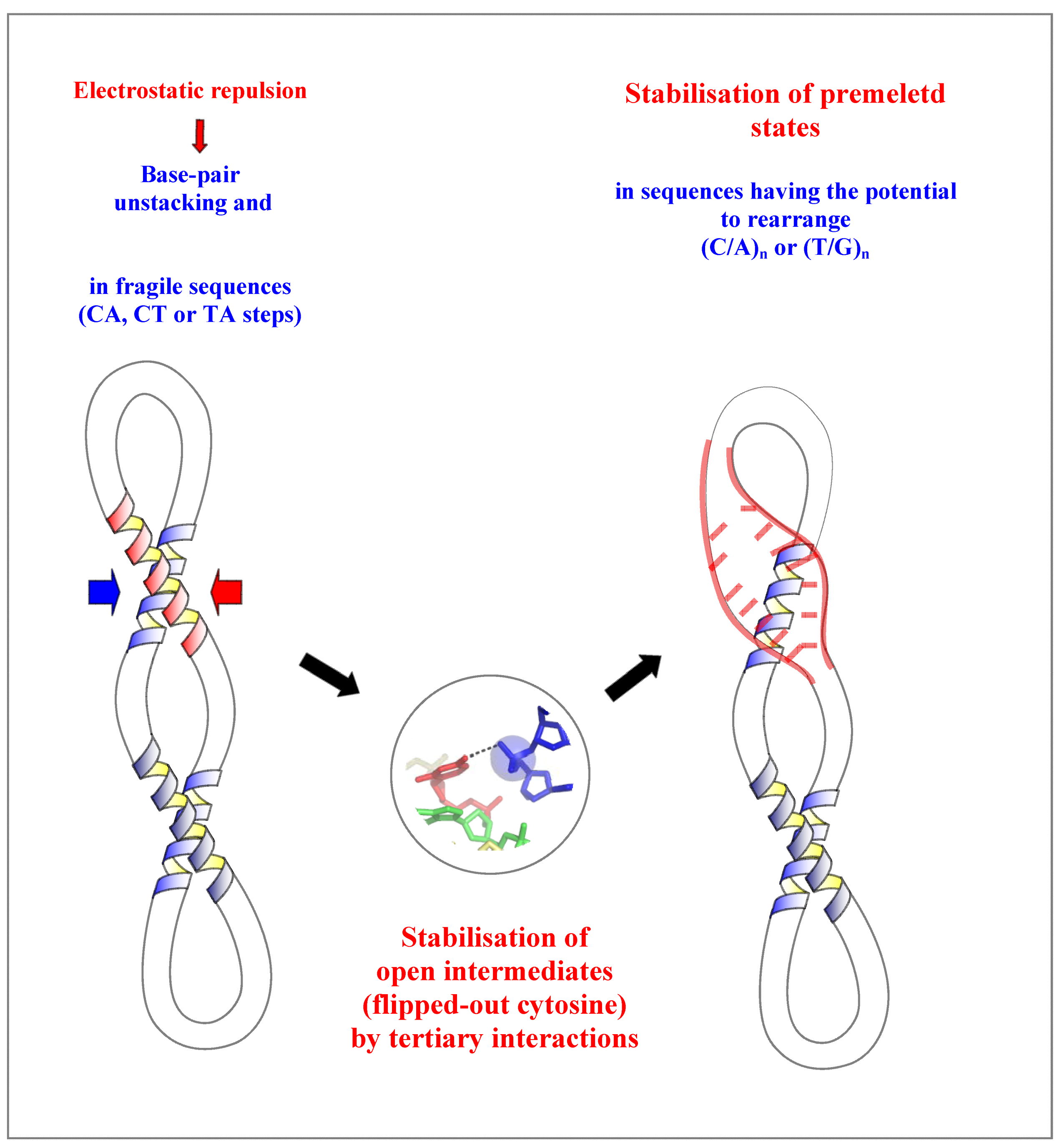
3. Conclusions
References
- Duderstadt, K.E.; Chuang, K.; Berger, J. DNA stretching by bacterial initiatorspromotes replication origin opening. Nature 2011, 478, 209–213. [Google Scholar] [CrossRef]
- Feklistov, A.; Darst, S.A. Structural basis for promoter −10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 2011, 147, 1257–1269. [Google Scholar] [CrossRef]
- Büttner, K.; Nehring, S.; Hopfner, K.-P. Structural basis for duplex separation by superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007, 14, 647–652. [Google Scholar] [CrossRef]
- Tuteja, N.; Tuteja, R. Unraveling DNA helicases. Motif, structure mechanism and function. Eur. J. Biochem. 2004, 271, 1849–1863. [Google Scholar]
- Travers, A.A. The structural basis of DNA flexibility. Phil. Trans. R. Soc. Lond. A 2004, 362, 1423–1438. [Google Scholar] [CrossRef]
- Lankas, F.; Sponer, J.; Langowski, J.; Cheatham, T. DNA base pair step deformability inferred from molecular dynamics simulation. Biophys. J. 2003, 85, 2872–2883. [Google Scholar] [CrossRef]
- Yakovchuk, P.; Protozanova, E.; Frank-Kamenetskii, M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef]
- Benham, C.J.; Mielke, S.P. DNA mechanics. Annu. Rev. Biomed. Eng. 2005, 7, 21–53. [Google Scholar] [CrossRef]
- Travers, A.; Muskhelishvili, G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007, 8, 147–151. [Google Scholar] [CrossRef]
- Echols, H. Nucleoprotein Structures Initiating DNA-Replication, Transcription, and Site-Specific Recombination. J. Biol. Chem. 1990, 265, 14697–14700. [Google Scholar]
- Grosschedl, R. Higher-Order Nucleoprotein Complexes in Transcription: Analogies with Site-Specific Recombination. Curr. Opin. Cell Biol. 1995, 7, 362–370. [Google Scholar] [CrossRef]
- Segal, E.; Widom, J. What controls nucleosome positions? Trends Genet. 2009, 25, 335–343. [Google Scholar] [CrossRef]
- Timsit, Y.; Moras, D. Cruciform structures and functions. Q. Rev. Biophys. 1996, 29, 279–307. [Google Scholar] [CrossRef]
- Timsit, Y.; Vilbois, E.; Moras, D. Base pairing shift in the major groove of (CA)n tracts byB-DNA crystal structures. Nature 1991, 354, 167–170. [Google Scholar] [CrossRef]
- Timsit, Y.; Moras, D. Self-fitting and self-modifying properties of the B-DNA molecule. J. Mol. Biol. 1995, 251, 629–647. [Google Scholar] [CrossRef]
- Várnai, P.; Timsit, Y. Differential stability of DNA crossovers in solution mediated by divalent cations. Nucleic Acid Res. 2010, 38, 4163–4172. [Google Scholar] [CrossRef]
- Timsit, Y.; Várnai, P. Helical chirality: A link between local interactions and global topology in DNA. PLoS One 2010, 5, e9326. [Google Scholar] [CrossRef]
- Timsit, Y. Local sensing of global DNA topology: From crossover geometry to type II topoisomerase processivity. Nucleic Acids Res. 2011, 39, 8665–8676. [Google Scholar] [CrossRef]
- Srinivasan, A.R.; Olson, W.K. DNA associations: Packing calculation in A-, B- and Z-DNA structures. Biophys. Chem. 1992, 43, 279–310. [Google Scholar] [CrossRef]
- Murthy, V.L.; Rose, G.D. Is counterion delocalization responsible for collapse in RNA folding? Biochemistry 2000, 39, 14365–14370. [Google Scholar]
- Timsit, Y.; Moras, D. [20] Crystallization of DNA. Meth. Enzymol. 1992, 211, 409–429. [Google Scholar] [CrossRef]
- Lu, X.-J.; Shakked, Z.; Olson, W. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000, 300, 819–840. [Google Scholar] [CrossRef]
- Timsit, Y.; Moras, D. Groove-Backbone Interaction in B-DNA: Implication for DNA condensation and recombination. J. Mol. Biol. 1991, 221, 919–940. [Google Scholar] [CrossRef]
- Timsit, Y.; Moras, D. DNA self-fitting: The double helix directs the geometry of its supramolecular assemblies. EMBO J. 1994, 13, 2737–2746. [Google Scholar]
- Timsit, Y.; Shatzky-Schwartz, M.; Shakked, Z. Left-handed DNA crossovers. Implications for DNA-DNA recognition and structural alterations. J. Biomol. Struct. Dyn. 1999, 16, 775–785. [Google Scholar]
- Nissen, P.; Ippolito, J.A.; Ban, N.; Moore, P.; Steitz, T. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA 2001, 98, 4899–4903. [Google Scholar] [CrossRef]
- Baeyens, K.; de Bondt, H.L.; Holbrook, S.R. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nat. Struct. Biol. 1996, 2, 56–62. [Google Scholar]
- Schindelin, H.; Zhang, M.; Bald, R.; Fürste, J.-P.; Erdmann, V.A.; Heinemann, U. Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 1995, 249, 595–603. [Google Scholar] [CrossRef]
- Gagnon, M.G.; Mukhopadhyay, A.; Steinberg, S.V. Close packing of helices 3 and 12 of 16S rRNA is required for the normal ribosome function. J. Biol. Chem. 2006, 281, 39349–39357. [Google Scholar] [CrossRef]
- Timsit, Y.; Westhof, E.; Fuchs, R.; Moras, D. Unusual helical packing in crystals of DNA bearing a mutation hot spot. Nature 1989, 341, 459–462. [Google Scholar] [CrossRef]
- Timsit, Y.; Varnai, P. Cytosine, the double helix and DNA self-assembly. J. Mol. Recognit. 2011, 24, 137–138. [Google Scholar] [CrossRef]
- Mayer-Jung, C.; Moras, D.; Timsit, Y. Effect of cytosine methylation on DNA-DNA recognition at CpG steps. J. Mol. Biol. 1997, 270, 328–335. [Google Scholar] [CrossRef]
- Qiu, X.; Andresen, K.; Kwok, L.W.; Lamb, J.S.; Park, H.Y.; Pollack, L. Inter-DNA attraction mediated by divalent counterions. Phys. Rev. Lett. 2007, 99, 038104–038107. [Google Scholar] [CrossRef]
- Tan, Z.J.; Chen, S.J. Electrostatic free energy landscapes for nucleic acid helix assembly. Nucleic Acids Res. 2006, 34, 6629–6639. [Google Scholar] [CrossRef]
- Inoue, S.; Sugiyama, S.; Travers, A.A.; Ohyama, T. Self-assembly of double-stranded DNA molecules at nanomolar concentrations. Biochemistry 2007, 46, 164–171. [Google Scholar]
- Ortiz-Lombardia, M.; Gonzalez, A.; Eritja, R.; Aymami, J.; Azorin, F.; Coll, M. Crystal structure of a DNA Holliday junction. Nat. Struct. Biol. 1999, 6, 913–917. [Google Scholar] [CrossRef]
- van Buuren, B.N.M.; Hermann, T.; Wijmenga, S.S.; Westhof, E. Brownian-dynamics simulations of metal-ion binding to four-way junctions. Nucleic Acids Res. 2002, 30, 507–514. [Google Scholar] [CrossRef]
- Woodson, S.A. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005, 9, 104–109. [Google Scholar] [CrossRef]
- Bloomfield, V.A. DNA condensation. Curr. Opin. Struct. Biol. 1996, 6, 334–341. [Google Scholar] [CrossRef]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. 2002, 3, 430–440. [Google Scholar] [CrossRef]
- Adrian, M.; Heggeler-Bordier, B.; Wahli, W.; Stasiak, A.Z.; Stasiak, A; Dubochet, J. Direct visualization of supercoiled DNA molecules in solution. EMBO J. 1990, 9, 4551–4554. [Google Scholar]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl. Acad. Sci. USA 1997, 94, 496–501. [Google Scholar] [CrossRef]
- Shlyakhtenko, L.S.; Miloseska, L.; Potaman, V.N.; Sinden, R.R.; Lyubchenko, Y.L. Intersegmental interactions in supercoiled DNA: Atomic force microscope study. Ultramicroscopy 2003, 97, 263–270. [Google Scholar] [CrossRef]
- Schlick, T.; Olson, W.K. Trefoil Knotting Revealed by Molecular-Dynamics Simulations of Supercoiled DNA. Science 1992, 257, 1110–1115. [Google Scholar]
- Robinson, P.J.J.; Rhodes, D. Structure of the ‘30 nm’ chromatin fibre: A key role for the linker histone. Curr. Opin. Struct. Biol. 2006, 16, 336–343. [Google Scholar] [CrossRef]
- Schalch, T.; Duda, S.; Sargent, D.F.; Richmond, T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 2005, 436, 138–141. [Google Scholar] [CrossRef]
- Wong, H.; Victor, J.M.; Mozziconacci, J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS One 2007, 2, e877. [Google Scholar] [CrossRef]
- Dickerson, R.E.; Goodsell, D.S.; Kopka, M.L.; Pjura, P.E. The effect of crystalpacking on oligonucleotide double helix structure. J. Biomol. Struct. Dyn. 1987, 5, 557–579. [Google Scholar] [CrossRef]
- Hays, F.A.; Teegarden, A.; Jones, Z.J.R.; Harms, M.; Raup, D.; Watson, J.; Vavaliere, E.; Ho, P.S. How sequence defines structure: A crystallographic map of DNA structure and conformation. Proc. Natl. Acad. Sci. USA 2005, 102, 7157–7162. [Google Scholar]
- van Aalten, D.M.; Erlanson, D.A.; Verdine, G.L.; Joshua-Tor, L. A structural snapshot of base-pair opening in DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 11809–11814. [Google Scholar]
- Hays, F.A.; Jones, Z.R.J.; Ho, P.S. Influence of minor groove substituents on the structure of DNA Holliday junctions. Biochemistry 2004, 43, 9813–9822. [Google Scholar] [CrossRef]
- Tari, L.W.; Secco, A.S. Base-pair opening and spermine binding —B-DNA features displayed in the crystal structure of a gal operon fragment: Implications for protein-DNA recognition. Nucleic Acids Res. 1995, 23, 2065–2073. [Google Scholar] [CrossRef]
- Hays, F.A.; Vargason, J.M.; Ho, P.S. Effect of sequence on the conformation of DNA Holliday junction. Biochemistry 2003, 42, 9586–9597. [Google Scholar] [CrossRef]
- Olson, W.K.; Gorin, A.A.; Lu, X.-J.; Hock, L.M.; Zhurkin, V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA 1998, 95, 11163–11168. [Google Scholar] [CrossRef]
- Wong, K.Y.; Pettitt, M. The pathway of oligomeric melting investigated by molecular dynamics simulation. Biophys. J. 2008, 95, 5618–5626. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Timsit, Y. DNA-Directed Base Pair Opening. Molecules 2012, 17, 11947-11964. https://doi.org/10.3390/molecules171011947
Timsit Y. DNA-Directed Base Pair Opening. Molecules. 2012; 17(10):11947-11964. https://doi.org/10.3390/molecules171011947
Chicago/Turabian StyleTimsit, Youri. 2012. "DNA-Directed Base Pair Opening" Molecules 17, no. 10: 11947-11964. https://doi.org/10.3390/molecules171011947




