Interaction of α-Melanocortin and Its Pentapeptide Antisense LVKAT: Effects on Hepatoprotection in Male CBA Mice
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Recognition of Amino Acids and Related Antisense Peptides
| Amino acid | Codons for amino acids | Kyte-Doolittle hydropathy scale | Antisense | ||
|---|---|---|---|---|---|
| subgroup | value | 3'→5' | 5'→3' | ||
| R (arginine) | cgc, cga, cgg, cgu, aga, agg | polar | −4.5 | A, S* | A, S*, P*, T* |
| K (lysine) | aaa, aag | polar | −3.9 | F | F, L |
| Q (glutamine) | caa, cag | polar | −3.5 | V | L |
| N (asparagine) | aac, aau | polar | −3.5 | L | I, V |
| E (glutamic acid) | gag, gaa | polar | −3.5 | L | L, F |
| D (aspartic acid) | gac, gau | polar | −3.5 | L | I, V |
| H (histidine) | cac, cau | polar | −3.2 | V | V, M |
| P (proline) | ccc, cca, ccu, ccg | neutral | −1.6 | G | G, W, R* |
| Y (tyrosine) | uac, uau | neutral | −1.3 | M*, I* | I*, V* |
| W (tryptophan) | ugg | neutral | −0.9 | T | P |
| S (serine) | ucg, uca, agc, agu, ucu, ucc | neutral | −0.8 | S, R* | G, R*, T, A* |
| T (threonine) | aca, acg, acc | neutral | −0.7 | W, C* | G, S, C*, R* |
| G (glycine) | ggg, ggu, gga, ggc | neutral | −0.4 | P | P, S, T, A* |
| A (alanine) | gcg, gcu, gcc, gca | nonpolar | 1.8 | R | R, G*, S*, C* |
| M (methionine) | aug | nonpolar | 1.9 | Y* | H |
| C (cysteine) | ugu, ugc | nonpolar | 2.5 | T* | T*, A* |
| F (phenylalanine) | uuu, uuc | nonpolar | 2.8 | K | K, E |
| L (leucine) | uug, uua, cuc, cuu, cug, cua | nonpolar | 3.8 | D, E, N | E, Q, K |
| V (valine) | guu, guc, gug, gua | nonpolar | 4.2 | H, Q | H, D, N, Y* |
| I (isoleucine) | aua | nonpolar | 4.5 | Y* | N, D, Y* |

2.2. Tryptophan Fluorescence Reveals Sense-Antisense Peptide Interaction



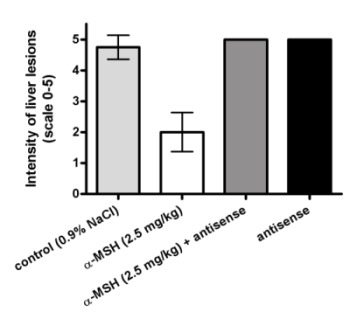
2.3. Modulation of α-MSH Hepatoprotection with Antisense Pentapeptide



3. Experimental
3.1. Test Compounds
3.2. Tryptophan Fluorescence Experiment
3.3. Treatment Regimen (Hepatotoxicity Model)
3.4. Histopathological and Transaminase Estimation of Liver Damage
3.5. Data Analysis
4. Conclusions
- (1) Transcription of α-MSH sequence in 3’→5’ direction was used to design antisense peptide (LVKAT) to the central region of α-MSH that serves as a pharmacophore for melanocortin 1, 3, 4 and 5 receptors.
- (2) Tryptophan fluorescence titration is a simple and efficient method to evaluate the binding of antisense peptide LVKAT to the α-MSH molecule in vitro.
- (3) Antisense peptide LVKAT abolished hepatoprotective effects of α-MSH in vivo.
Acknowledgments
Conflict of Interest
- Samples Availability: Not available.
References
- Blalock, J.E.; Bost, K.L. Binding of peptides that are specified by complementary RNAs. Biochem. J. 1986, 234, 679–683. [Google Scholar]
- Biro, J.C. The proteonomic code: A molecular recognition code for proteins. Theor. Biol. Med. Model. 2007, 4, 1–45. [Google Scholar] [CrossRef]
- Heal, J.R.; Roberts, G.W.; Raynes, J.G.; Bhakoo, A.; Miller, A.D. Specific Interactions between sense and complementary peptides: the basis for the proteomic code. ChemBiochem 2002, 3, 136–151. [Google Scholar] [CrossRef]
- Blalock, J.E. Genetic origin of protein shape and interaction rules. Nat. Med. 1995, 1, 876–878. [Google Scholar] [CrossRef]
- Štambuk, N.; Konjevoda, P.; Boban-Blagaić, A.; Pokrić, B. Molecular recognition theory of the complementary (antisense) peptide interactions. Theory Biosci. 2005, 123, 265–275. [Google Scholar] [CrossRef]
- McGuire, K.L.; Holmes, D.S. Role of complementary proteins in autoimmunity: an old idea re-emerges with new twists. Trends Immunol. 2005, 26, 367–372. [Google Scholar] [CrossRef]
- Blalock, J.E.; Smith, E.M. Hydropathic anti-complementarity of amino acids based on the genetic code. Biochem. Biophys. Res. Commun. 1984, 121, 203–207. [Google Scholar] [CrossRef]
- Baranyi, L.; Campbell, W.; Ohshima, K.; Fujimoto, S.; Boros, M.; Okada, H. The antisense homology box: A new motif within proteins that encodes biologically active peptides. Nat. Med. 1995, 1, 894–901. [Google Scholar] [CrossRef]
- Heal, J.R.; Roberts, G.W.; Christie, G.; Miller, A.D. Inhibition of β-amyloid aggregation and neurotoxicity by complementary (antisense) peptides. ChemBiochem 2002, 3, 86–92. [Google Scholar] [CrossRef]
- Heal, J.R.; Bino, S.; Roberts, G.W.; Raynes, J.G.; Miller, A.D. Mechanistic Investigation into Complementary (Antisense) Peptide Mini-Receptor Inhibitors of Cytokine Interleukin-1. ChemBiochem 2002, 3, 76–85. [Google Scholar] [CrossRef]
- Bost, K.L.; Blalock, E. Complementary peptides as interactive sites for protein binding. Viral Immunol. 1989, 2, 229–238. [Google Scholar] [CrossRef]
- Lipton, J.M.; Catania, A. Anti-inflammatory actions of the neuroimmunomodulator alpha-MSH. Immunol. Today 1997, 18, 140–145. [Google Scholar] [CrossRef]
- Brzoska, T.; Luger, T.A.; Maaser, C.; Abels, C.; Böhm, M. Alpha-melanocyte-stimulating hormone and related tripeptides: biochemistry, antiinflammatory and protective effects in vitro and in vivo, and future perspectives for the treatment of immune-mediated inflammatory diseases. Endocr. Rev. 2008, 29, 581–602. [Google Scholar] [CrossRef]
- Getting, S.J. Targeting melanocortin receptors as potential novel therapeutics. Pharmacol. Ther. 2006, 111, 1–15. [Google Scholar] [CrossRef]
- Turčić, P.; Bradamante, M.; Houra, K.; Štambuk, N.; Kelava, T.; Konjevoda, P.; Kazazić, S.; Vikić-Topić, D.; Pokrić, B. Effects of α-Melanocortin Enantiomers on Acetaminophen-Induced Hepatotoxicity in CBA Mice. Molecules 2009, 14, 5017–5026. [Google Scholar] [CrossRef]
- Blagaić, V.; Houra, K.; Turčić, P.; Štambuk, N.; Konjevoda, P.; Boban-Blagaić, A.; Kelava, T.; Kos, M.; Aralica, G.; Čulo, F. The Influence of α-, β-, and γ-Melanocyte Stimulating Hormone on Acetaminophen Induced Liver Lesions in Male CBA Mice. Molecules 2010, 15, 1232–1241. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S. Amino acid pairing. J. Theor. Biol. 1982, 94, 885–894. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S. Peptide self-aggregation and peptide complementarity as bases for the evolution of peptide receptors: A review. J. Mol. Recognit. 2005, 18, 40–49. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S.; Westall, F.C. Bovine pineal antireproductive tripeptide binds to luteinizing hormone-releasing hormone: A model for peptide modulation by sequence specific peptide interactions? Brain Res. Bull. 1986, 17, 519–528. [Google Scholar] [CrossRef]
- Štambuk, N. On the genetic origin of complementary protein coding. Croat. Chem. Acta 1998, 71, 573–589. [Google Scholar]
- Houra, K.; Štambuk, N.; Konjevoda, P.; Boban-Blagaić, A.; Bruketa, T.; Pokrić, B. Alpha-melanotropin peptide: Structure and ligand-receptor recognition. Croat. Chem. Acta 2006, 79, 379–383. [Google Scholar]
- Štambuk, N.; Konjevoda, P.; Vikić-Topić, D.; Pokrić, B. Modelling of ICAM-1 and LFA-1 interaction using molecular recognition theory. Croat. Chem. Acta 2008, 81, 283–287. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T. Paleontological Data Analysis; Blackwell: Oxford, UK, 2006; pp. 67–75. [Google Scholar]
- Gampp, H.; Maeder, M.; Meyer, C.J.; Zuberbühler, A.D. Calculation of equilibrium constants from multiwavelength spectroscopic data-I Mathematical considerations. Talanta 1985, 32, 95–101. [Google Scholar] [CrossRef]
- Gampp, H.; Maeder, M.; Meyer, C.J.; Zuberbühler, A.D. Calculation of equilibrium constants from multiwavelength spectroscopic data-II: SPECFIT: Two user-friendly programs in basic and standard FORTRAN 77. Talanta 1985, 32, 257–264. [Google Scholar] [CrossRef]
- Gampp, H.; Maeder, M.; Meyer, C.J.; Zuberbühler, A.D. Calculation of equilibrium constants from multiwavelength spectroscopic data-IV Model-free least-squares refinement by use of evolving factor analysis. Talanta 1986, 33, 943–951. [Google Scholar] [CrossRef]
- Maeder, M.; Neuhold, Y.-M. Practical Data Analysis, 1st ed; Elsevier B. V.: Amsterdam, The Netherlands, 2007; pp. 214–217. [Google Scholar]
- Bost, K.L.; Smith, E.M.; Blalock, J.E. Similarity between the corticotropin (ACTH) receptor and a peptide encoded by an RNA that is complementary to ACTH mRNA. Proc. Natl. Acad. Sci. USA 1985, 82, 1372–1375. [Google Scholar] [CrossRef]
- Fassina, G.; Cassini, G. Design and recognition properties of a hydropathically complementary peptide to human interleukin 1β. Biochem. J. 1992, 282, 773–779. [Google Scholar]
- Loo, J.L. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997, 16, 1–23. [Google Scholar] [CrossRef]
- Bhakoo, A.; Raynes, J.G.; Heal, J.R.; Keller, M.K.; Miller, A.D. De-novo design of complementary (antisense) peptide mini-receptor inhibitor of interleukin 18 (IL-18). Mol. Immunol. 2004, 41, 1217–1224. [Google Scholar] [CrossRef]
- Silva, V.M.; Chen, C.; Hennig, G.E.; Whiteley, H.E.; Manautou, E.J. Changes in susceptibility to acetaminophen-induced liver injury by the organic anion indocyanine green. Food. Chem. Toxicol. 2001, 3, 271–278. [Google Scholar]
- Ping-Lim, S.; Andrews, F.J.; O'Brien, P.E. Misoprostol protection against acetaminophen-induced hepatotoxicity in the rat. Dig. Dis. Sci. 1994, 39, 1249–1256. [Google Scholar] [CrossRef]
- Newsome, P.N.; Plevris, J.N.; Nelson, L.N.; Hayes, P.C. Animal models of fulminant hepatic failure: A critical evaluation. Liver Transpl. 2000, 6, 21–31. [Google Scholar]
- Kluczyk, A.; Cebrat, M.; Zbozień-Pacamaj, R.; Lisowski, M.; Stefanowicz, P.; Wieczorek, Z.; Siemion, I.Z. On the peptide-antipeptide interactions in interleukin-1 receptor system. Acta Biochem. Pol. 2004, 51, 57–66. [Google Scholar]
- Siemion, I.Z.; Zbozien-Pacamaj, R; Stefanowicz, P. New hypothesis on amino acid complementarity and its evaluation on TGF-β2-related peptides. J. Mol. Recognit. 2001, 14, 1–12. [Google Scholar] [CrossRef]
- Guarner, F.; Boughton-Smith, N.K.; Blackwell, G.J.; Moncada, S. Reduction by prostacyclin of acetaminophen-induced liver toxicity in the mouse. Hepatology 1988, 8, 248–253. [Google Scholar] [CrossRef]
- Čulo, F.; Renić, M.; Sabolović, D.; Radoš, M.; Bilić, A.; Jagić, V. Ketoconazole inhibits acetaminophen-induced hepatotoxicity in mice. Eur. J. Gastroenterol. Hepatol. 1995, 7, 757–762. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Houra, K.; Turčić, P.; Gabričević, M.; Weitner, T.; Konjevoda, P.; Štambuk, N. Interaction of α-Melanocortin and Its Pentapeptide Antisense LVKAT: Effects on Hepatoprotection in Male CBA Mice. Molecules 2011, 16, 7331-7343. https://doi.org/10.3390/molecules16097331
Houra K, Turčić P, Gabričević M, Weitner T, Konjevoda P, Štambuk N. Interaction of α-Melanocortin and Its Pentapeptide Antisense LVKAT: Effects on Hepatoprotection in Male CBA Mice. Molecules. 2011; 16(9):7331-7343. https://doi.org/10.3390/molecules16097331
Chicago/Turabian StyleHoura, Karlo, Petra Turčić, Mario Gabričević, Tin Weitner, Paško Konjevoda, and Nikola Štambuk. 2011. "Interaction of α-Melanocortin and Its Pentapeptide Antisense LVKAT: Effects on Hepatoprotection in Male CBA Mice" Molecules 16, no. 9: 7331-7343. https://doi.org/10.3390/molecules16097331