Modulation of the Pharmacological Activities of Secretory Phospholipase A2 from Crotalus durissus cascavella Induced by Naringin
Abstract
:1. Introduction
2. Results and Discussion
2.1. Enzymatic activity
2.2. Pharmacological properties
2.3. Chemical and structural modifications
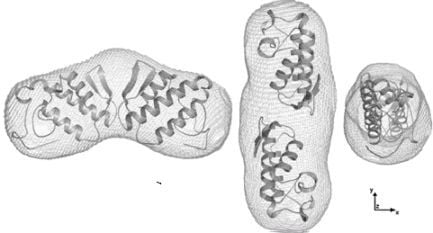
2.4. Dimer models in solution for sPLA2 from C. d. cascavella
3. Experimental
3.1. Venom, animals and reagents
3.2. Purification of sPLA2 from Crotalus durissus cascavella venom
3.3. Incubation of sPLA2 with naringin and purification of modified sPLA2
3.4. Measurement of sPLA2 oxidation
3.5. Measurement of sPLA2 activity
3.6. Myotoxic activity
3.7. Neurotoxic effect assay
3.8. Platelet aggregation studies
3.9. Paw edema assay
3.10. Antibacterial activity
3.11. Circular dichroism (CD) spectroscopy
3.12. Mass spectroscopy
3.13. Small angle X-ray scattering (SAXS)
3.14. Additional computational analysis
3.15. Statistical analyses
4. Conclusions
Acknowledgements
References and Notes
- Di Carlo, G.; Mascolo, N.; Izzo, A.; Capasso, F. Flavonoids: Old and new aspects of a class of natural therapeutic drugs. Life Sci. 1999, 65, 337–353. [Google Scholar] [CrossRef]
- Fawzy, A.; Vishwanath, B.; Franson, R. Inhibition of human non-pancreatic phospholipases A2 by retinoids and flavonoids. Mechanism of action. Agents Actions 1988, 25, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, B.; Fawzy, A.; Franson, R. Edema-inducing activity of phospholipase A2 purified from human synovial fluid and inhibition by aristolochic acid. Inflammation 1988, 12, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Guardia, T.; Rotelli, A.; Juarez, A.; Pelzer, L. Anti-inflammatory properties of plant flavonoids. effects of rutin, quercetin and hesperidin on adjuvant arthritis in rat. Farmaco 2001, 56, 683–687. [Google Scholar] [CrossRef]
- Lindahl, M.; Tagesson, C. Flavonoids as phospholipase A2 inhibitors: Importance of their structure for selective inhibition of group ii phospholipase A2. Inflammation 1997, 21, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Rotelli, A.; Guardia, T.; Juárez, A.; de la Rocha, N.; Pelzer, L. Comparative study of flavonoids in experimental models of inflammation. Pharmacol. Res. 2003, 48, 601–606. [Google Scholar] [CrossRef]
- Iglesias, C.; Aparicio, R.; Rodrigues-Simioni, L.; Camargo, E.; Antunes, E.; Marangoni, S.; de Oliveira Toyama, D.; Beriam, L.; Monteiro, H.; Toyama, M. Effects of morin on snake venom phospholipase A2 (PLA2). Toxicon 2005, 46, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Toyama, D.; Marangoni, S.; Diz-Filho, E.; Oliveira, S.; Toyama, M. Effect of umbelliferone (7-hydroxycoumarin, 7-hoc) on the enzymatic, edematogenic and necrotic activities of secretory phospholipase A2 (sPLA2) isolated from Crotalus durissus collilineatus venom. Toxicon 2009, 53, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Cirino, G. Multiple controls in inflammation. Extracellular and intracellular phospholipase A2, inducible and constitutive cyclooxygenase, and inducible nitric oxide synthase. Biochem. Pharmacol. 1998, 55, 105–111. [Google Scholar] [CrossRef]
- Fuentes, L.; Hernández, M.; Nieto, M.; Sánchez Crespo, M. Biological effects of group iia secreted phosholipase a(2). FEBS Lett. 2002, 531, 7–11. [Google Scholar] [CrossRef]
- Gil, B.; Sanz, M.; Terencio, M.; Gunasegaran, R.; Payá, M.; Alcaraz, M. Morelloflavone, a novel biflavonoid inhibitor of human secretory phospholipase A2 with anti-inflammatory activity. Biochem. Pharmacol. 1997, 53, 733–740. [Google Scholar] [CrossRef]
- Câmara, P.; Esquisatto, L.; Camargo, E.; Ribela, M.; Toyama, M.; Marangoni, S.; De Nucci, G.; Antunes, E. Inflammatory edema induced by phospholipases A2 isolated from Crotalus durissus sp. in the rat dorsal skin: A role for mast cells and sensory c-fibers. Toxicon 2003, 41, 823–829. [Google Scholar] [CrossRef]
- Lee, W.; Toyama, M.; Soares, A.; Giglio, J.; Marangoni, S.; Polikarpov, I. Crystallization and preliminary X-ray diffraction studies of piratoxin iii, a d-49 phospholipase A2 from the venom of Bothrops pirajai. Acta. Crystallogr. D. Biol. Crystallogr. 1999, 55, 1229–1230. [Google Scholar] [PubMed]
- Benavente-García, O.; Castillo, J. Update on uses and properties of citrus flavonoids: New findings in anticancer, cardiovascular, and anti-inflammatory activity. J. Agric. Food. Chem. 2008, 56, 6185–6205. [Google Scholar] [CrossRef] [PubMed]
- Hodek, P.; Trefil, P.; Stiborová, M. Flavonoids-potent and versatile biologically active compounds interacting with cytochromes p450. Chem. Biol. Interact. 2002, 139, 1–21. [Google Scholar] [CrossRef]
- Benavente-García, O.; Castillo, J.; Marin, F.R.; Ortuño, A.; Del Río, J.A. Uses and properties of citrus flavonoids. J. Agric. Food. Chem. 1997, 45, 11. [Google Scholar] [CrossRef]
- Svergun, D.; Koch, M. Advances in structure analysis using small-angle scattering in solution. Curr. Opin. Struct. Biol. 2002, 12, 654–660. [Google Scholar] [CrossRef]
- Arni, R.; Ward, R. Phospholipase A2 a structural review. Toxicon 1996, 34, 827–841. [Google Scholar] [CrossRef]
- Burke, J.; Dennis, E. Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid. Res. 2009, 50, S237–S242. [Google Scholar] [CrossRef] [PubMed]
- Arni, R.; Fontes, M.; Barberato, C.; Gutiérrez, J.; Díaz, C.; Ward, R. Crystal structure of myotoxin ii, a monomeric lys49-phospholipase A2 homologue isolated from the venom of Cerrophidion (Bothrops) godmani. Arch. Biochem. Biophys. 1999, 366, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Viçoti, M.; Abrego, J.; Lourenzoni, M.; Cintra, A.; Arruda, E.; Tomaz, M.; Melo, P.; Arni, R. Interfacial surface charge and free accessibility to the PLA2-active site-like region are essential requirements for the activity of lys49 PLA2 homologues. Toxicon 2007, 49, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Arni, R.; Ward, R.; Gutierrez, J.; Tulinsky, A. Structure of a calcium-independent phospholipase-like myotoxic protein from Bothrops asper venom. Acta. Crystallogr. D. Biol. Crystallogr. 1995, 51, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, M.; Fagundes, F.; Teixeira, B.; Toyama, M.; Aparicio, R. Purification and preliminary crystallographic analysis of a new lys49-pla2 from B. jararacussu. Int. J. Mol. Sci. 2008, 9, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.; de Azevedo, W.J.; Arni, R. At the interface: crystal structures of phospholipases A2. Toxicon 1998, 36, 1623–1633. [Google Scholar] [CrossRef]
- da Silva Giotto, M.; Garratt, R.; Oliva, G.; Mascarenhas, Y.; Giglio, J.; Cintra, A.; de Azevedo, W.J.; Arni, R.; Ward, R. Crystallographic and spectroscopic characterization of a molecular hinge: Conformational changes in bothropstoxin i, a dimeric lys49-phospholipase A2 homologue. Proteins 1998, 30, 442–454. [Google Scholar] [CrossRef]
- Marchi-Salvador, D.; Corrêa, L.; Magro, A.; Oliveira, C.; Soares, A.; Fontes, M. Insights into the role of oligomeric state on the biological activities of crotoxin: crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin b from Crotalus durissus terrificus venom. Proteins 2008, 72, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zhou, Y.; Lin, Z. Structure of agkistrodotoxin in an orthorhombic crystal form with six molecules per asymmetric unit. Acta. Crystallogr. D. Biol. Crystallogr. 1999, 55, 1986–1996. [Google Scholar] [CrossRef] [PubMed]
- Magro, A.; Takeda, A.; Soares, A.; Fontes, M. Structure of btha-i complexed with p-bromophenacyl bromide: possible correlations with lack of pharmacological activity. Acta. Crystallogr. D. Biol. Crystallogr. 2005, 61, 1670–1677. [Google Scholar] [CrossRef] [PubMed]
- Boilard, E.; Rouault, M.; Surrel, F.; Le Calvez, C.; Bezzine, S.; Singer, A.; Gelb, M.; Lambeau, G. Secreted phospholipase A2 inhibitors are also potent blockers of binding to the m-type receptor. Biochemistry 2006, 45, 13203–13218. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, D.; Lopes-Ferreira, M.; Faquim-Mauro, E.; Macedo, M.; Farsky, S. Role of crotoxin, a phospholipase a2 isolated from Crotalus durissus terrificus snake venom, on inflammatory and immune reactions. Mediators. Inflamm. 2001, 10, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.J.; Kandaswami, C.; Theoharides, T. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Nevalainen, T.; Graham, G.; Scott, K. Antibacterial actions of secreted phospholipases A2. Review. Biochim. Biophys. Acta. 2008, 1781, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.; Sigler, P. Structure and catalytic mechanism of secretory phospholipases A2. Adv. Protein. Chem. 1994, 45, 53–88. [Google Scholar] [PubMed]Rigden, D.; Hwa, L.; Marangoni, S.; Toyama, M.; Polikarpov, I. The structure of the d49 phospholipase A2 piratoxin iii from Bothrops pirajai reveals unprecedented structural displacement of the calcium-binding loop: Possiblerelationship to cooperative substrate binding. Acta. Crystallogr. D. Biol. Crystallogr. 2003, 59, 255–262. [Google Scholar]
- Al-Lazikani, B.; Jung, J.; Xiang, Z.; Honig, B. Protein structure prediction. Curr. Op. Chem. Biol. 2001, 51–56. [Google Scholar] [CrossRef]
- Ponder, J.; Richards, F. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 1987, 193, 775–791. [Google Scholar] [CrossRef]
- Levitt, M.; Gerstein, M.; Huang, E.; Subbiah, S.; Tsai, J. Protein folding: the endgame. Ann. Rev. Biochem. 1997, 66, 549–579. [Google Scholar] [CrossRef] [PubMed]
- Toyama, M.; de Oliveira, D.; Beriam, L.; Novello, J.; Rodrigues-Simioni, L.; Marangoni, S. Structural, enzymatic and biological properties of new PLA(2) isoform from Crotalus durissus terrificus venom. Toxicon 2003, 41, 1033–1038. [Google Scholar] [CrossRef]
- Toyama, M.; Mancuso, L.; Giglio, J.; Novello, J.; Oliveira, B.; Marangoni, S. A quick procedure for the isolation of dimeric piratoxins-i and ii, two myotoxins from Bothrops pirajai snake venom. N-terminal sequencing. Biochem. Mol. Biol. Int. 1995, 37, 1047–1055. [Google Scholar] [PubMed]
- Soares, A.; Rodrigues, V.; Homsi-Brandeburgo, M.; Toyama, M.; Lombardi, F.; Arni, R.; Giglio, J. A rapid procedure for the isolation of the lys-49 myotoxin ii from Bothrops moojeni (caissaca) venom: biochemical characterization, crystallization, myotoxic and edematogenic activity. Toxicon. 1998, 36, 503–514. [Google Scholar] [CrossRef]
- Diz Filho, E.; Marangoni, S.; Toyama, D.; Fagundes, F.; Oliveira, S.; Fonseca, F.; Calgarotto, A.; Joazeiro, P.; Toyama, M. Enzymatic and structural characterization of new PLA2 isoform isolated from white venom of Crotalus durissus ruruima. Toxicon 2009, 53, 104–114. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, D.; Toyama, M.; Martins, A.; Havt, A.; Nobre, A.; Marangoni, S.; Câmara, P.; Antunes, E.; de Nucci, G.; Beliam, L.; Fonteles, M.; Monteiro, H. Structural and biological characterization of a crotapotin isoform isolated from Crotalus durissus cascavella venom. Toxicon 2003, 42, 53–62. [Google Scholar] [CrossRef]
- Toyama, M.; Toyama, D.; Joazeiro, P.; Carneiro, E.; Beriam, L.; Marangoni, L.; Boschero, A. Biological and structural characterization of a new PLA2 from the Crotalus durissus collilineatus venom. Protein J. 2005, 24, 103–112. [Google Scholar] [CrossRef] [PubMed]
- de Castro, R.; Landucci, E.; Toyama, M.; Giglio, J.; Marangoni, S.; De Nucci, G.; Antunes, E. Leucocyte recruitment induced by type ii phospholipases A(2) into the rat pleural cavity. Toxicon 2000, 38, 1773–1785. [Google Scholar] [CrossRef]
- Hernandez-Oliveira, S.; Toyama, M.; Toyama, D.; Marangoni, S.; Hyslop, S.; Rodrigues-Simioni, L. Biochemical, pharmacological and structural characterization of a new PLA2 from Crotalus durissus terrificus (south american rattlesnake) venom. Protein J. 2005, 24, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Rigden, D.; Hwa, L.; Marangoni, S.; Toyama, M.; Polikarpov, I. The structure of the d49 phospholipase A2 piratoxin iii from Bothrops pirajai reveals unprecedented structural displacement of the calcium-binding loop: Possiblerelationship to cooperative substrate binding. Acta. Crystallogr. D. Biol. Crystallogr. 2003, 59, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Ginsborg, B.; Warriner, J. The isolated chick biventer cervicis nerve-muscle preparation. Br. J. Pharmacol. Chemother. 1960, 15, 410–411. [Google Scholar] [CrossRef] [PubMed]
- Belo, C.; Leite, G.; Toyama, M.; Marangoni, S.; Corrado, A.; Fontana, M.; Southan, A.; Rowan, E.; Hyslop, S.; Rodrigues-Simioni, L. Pharmacological and structural characterization of a novel phospholipase A2 from Micrurus dumerilii carinicauda venom. Toxicon 2005, 46, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, F.; Antunes, E.; Morganti, R.; Monteiro, H.; Martins, A.; Toyama, D.; Marangoni, S.; Toyama, M. Characterization of a new platelet aggregating factor from crotoxin Crotalus durissus cascavella venom. Protein J. 2006, 25, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Santi-Gadelha, T.; Rocha, B.; Oliveira, C.; Aragão, K.; Marinho, E.; Gadelha, C.; Toyama, M.; Pinto, V.; Nagano, C.; Delatorre, P.; Martins, J.; Galvani, F.; Sampaio, A.; Debray, H.; Cavada, B. Purification of a pha-like chitin-binding protein from acacia farnesiana seeds: A time-dependent oligomerization protein. Appl. Biochem. Biotechnol. 2008, 150, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Toyama, M.; Toyama, D.O.; Passero, L.; Laurenti, M.; Corbett, C.; Tomokane, T.; Fonseca, F.; Antunes, E.; Joazeiro, P.; Beriam, L.; Martins, M.; Monteiro, H.; Fonteles, M. Isolation of a new l-amino acid oxidase from Crotalus durissus cascavella venom. Toxicon 2006, 47, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Kellermann, G.; Vicentin, F.; Tamura, E.; Rocha, M.; Tolentino, H.; Barbosa, A.; Craievich, A.; Torriani, I. The small-angle x-ray scattering beamline of the brazilian synchrotron light laboratory. J. Appl. Cryst. 1997, 30, 4. [Google Scholar] [CrossRef]
- Cavalcanti, L.; Torriani, I.; Plivelic, T.; Oliveira, C.; Kellermann, G.; Neuenschwander, R. Two new sealed sample cells for small angle x-ray scattering from macromolecules in solution and complex fluids using synchrotron radiation. Rev. Scient. Instrum. 2004, 4541–4546. [Google Scholar] [CrossRef]
- Svergun, D.; Koch, M. Small-angle scattering studies of biological macromolecules in solution. Rep. Progres. Physics. 2003, 1735–1782. [Google Scholar] [CrossRef]
- Tsutakawa, S.; Hura, G.; Frankel, K.; Cooper, P.; Tainer, J. Structural analysis of flexible proteins in solution by small angle x-ray scattering combined with crystallography. J. Struct. Biol. 2007, 158, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Hammersley, A.; Svensson, S.; Hanfland, M.; Fitch, A.; Hausermann, D. Two-dimensional detector software: From real detector to idealised image or two-theta scan. High. Press. Res. 1996, 235–248. [Google Scholar] [CrossRef]
- Konarev, P.; Volkov, V.; Sokolova, A.; Koch, M.; Svergun, D. Primus: A windows pc-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003, 1277–1282. [Google Scholar] [CrossRef]
- Svergun, D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992, 495–503. [Google Scholar] [CrossRef]
- Guinier, A.; Fournet, G. Small-Angle Scattering of x-Rays; John Wiley & Sons: New York, NY, USA, 1955. [Google Scholar]
- Semisotnov, G.; Kihara, H.; Kotova, N.; Kimura, K.; Amemiya, Y.; Wakabayashi, K.; Serdyuk, I.; Timchenko, A.; Chiba, K.; Nikaido, K.; Ikura, T.; Kuwajima, K. Protein globularization during folding. A study by synchrotron small-angle x-ray scattering. J. Mol. Biol. 1996, 262, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Putnam, C.; Hammel, M.; Hura, G.; Tainer, J. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007, 40, 191–285. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.; Oliveira Neto, M.D.; Napolitano, H.B.; Polikarpov, I.; Craievich, A.F. Determination of the molecular weight of proteins in solution from a single small-angle x-ray scattering measurement on a relative scale. J. Appl. Cryst. 2010, 43, 9. [Google Scholar] [CrossRef]
- Svergun, D.; Petoukhov, M.; Koch, M. Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 2001, 80, 2946–2953. [Google Scholar] [CrossRef]
- Volkov, V.; Svergun, D. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003, 860–864. [Google Scholar] [CrossRef]
- CCP4 High-throughput structure determination. In Proceedings of the 2002 ccp4 (collaborative computational project in macromolecular crystallography) study weekend, York, UK, 2002. Acta Crystallogr. D. Biol. Crystallogr. D58.
- Kozin, M.; Svergun, D. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001, 33–41. [Google Scholar] [CrossRef]
- Svergun, D.; Barberato, C.; Koch, M.H.J. Crysol - a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995, 28, 6. [Google Scholar] [CrossRef]
- Petoukhov, M.; Svergun, D. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005, 89, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Wriggers, W.; Milligan, R.; McCammon, J. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999, 125, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M. Swiss-model: an automated protein homology-modeling server. Nucleic. Acids. Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Jones, D. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, L.; Bryson, K.; Jones, D. The psipred protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Contact the authors. |








© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Santos, M.L.; Toyama, D.O.; Oliveira, S.C.B.; Cotrim, C.A.; Diz-Filho, E.B.S.; Fagundes, F.H.R.; Soares, V.C.G.; Aparicio, R.; Toyama, M.H. Modulation of the Pharmacological Activities of Secretory Phospholipase A2 from Crotalus durissus cascavella Induced by Naringin. Molecules 2011, 16, 738-761. https://doi.org/10.3390/molecules16010738
Santos ML, Toyama DO, Oliveira SCB, Cotrim CA, Diz-Filho EBS, Fagundes FHR, Soares VCG, Aparicio R, Toyama MH. Modulation of the Pharmacological Activities of Secretory Phospholipase A2 from Crotalus durissus cascavella Induced by Naringin. Molecules. 2011; 16(1):738-761. https://doi.org/10.3390/molecules16010738
Chicago/Turabian StyleSantos, Marcelo L., Daniela O. Toyama, Simone C. B. Oliveira, Camila A. Cotrim, Eduardo B. S. Diz-Filho, Fábio H. R. Fagundes, Veronica C. G. Soares, Ricardo Aparicio, and Marcos H. Toyama. 2011. "Modulation of the Pharmacological Activities of Secretory Phospholipase A2 from Crotalus durissus cascavella Induced by Naringin" Molecules 16, no. 1: 738-761. https://doi.org/10.3390/molecules16010738