1
Department of Experimental Medicine, University of Rome Tor Vergata, 00133 Rome, Italy
2
Department of Microbiology and Parasitology, Faculty of Pharmacy, Complutense University of Madrid, 28040 Madrid, Spain
3
Department of Pharmacy, Health and Nutritional Sciences, University of Calabria, 87036 Rende, Italy
Microorganisms 2021, 9(12), 2537; https://doi.org/10.3390/microorganisms9122537 - 8 Dec 2021
Cited by 11 | Viewed by 3660
Abstract
The chronic infection established by the human immunodeficiency virus 1 (HIV-1) produces serious CD4+ T cell immunodeficiency despite the decrease in HIV-1 ribonucleic acid (RNA) levels and the raised life expectancy of people living with HIV-1 (PLWH) through treatment with combined antiretroviral therapies
[...] Read more.
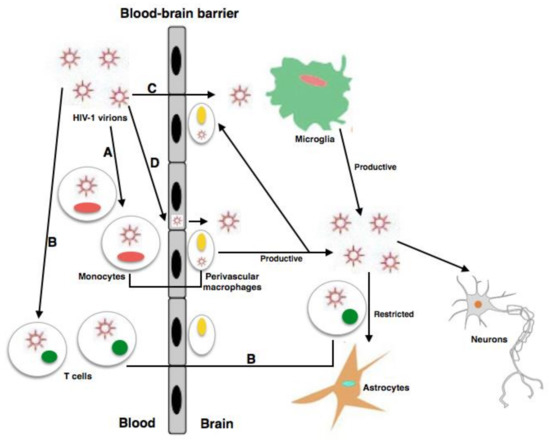
The chronic infection established by the human immunodeficiency virus 1 (HIV-1) produces serious CD4+ T cell immunodeficiency despite the decrease in HIV-1 ribonucleic acid (RNA) levels and the raised life expectancy of people living with HIV-1 (PLWH) through treatment with combined antiretroviral therapies (cART). HIV-1 enters the central nervous system (CNS), where perivascular macrophages and microglia are infected. Serious neurodegenerative symptoms related to HIV-associated neurocognitive disorders (HAND) are produced by infection of the CNS. Despite advances in the treatment of this infection, HAND significantly contribute to morbidity and mortality globally. The pathogenesis and the role of inflammation in HAND are still incompletely understood. Principally, growing evidence shows that the CNS is an anatomical reservoir for viral infection and replication, and that its compartmentalization can trigger the evolution of neurological damage and thus make virus eradication more difficult. In this review, important concepts for understanding HAND and neuropathogenesis as well as the viral proteins involved in the CNS as an anatomical reservoir for HIV infection are discussed. In addition, an overview of the recent advancements towards therapeutic strategies for the treatment of HAND is presented. Further neurological research is needed to address neurodegenerative difficulties in people living with HIV, specifically regarding CNS viral reservoirs and their effects on eradication.
Full article
(This article belongs to the Special Issue Molecular and Therapeutic Aspects of Viral Infections)
▼
Show Figures