Glycosylation of HIV Env Impacts IgG Subtype Responses to Vaccination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Expression Analyses
2.3. Mice and Immunization
2.4. Determination of Humoral Immune Responses
2.5. Determination of Cellular Immune Response
2.6. Statistical Analysis
3. Results
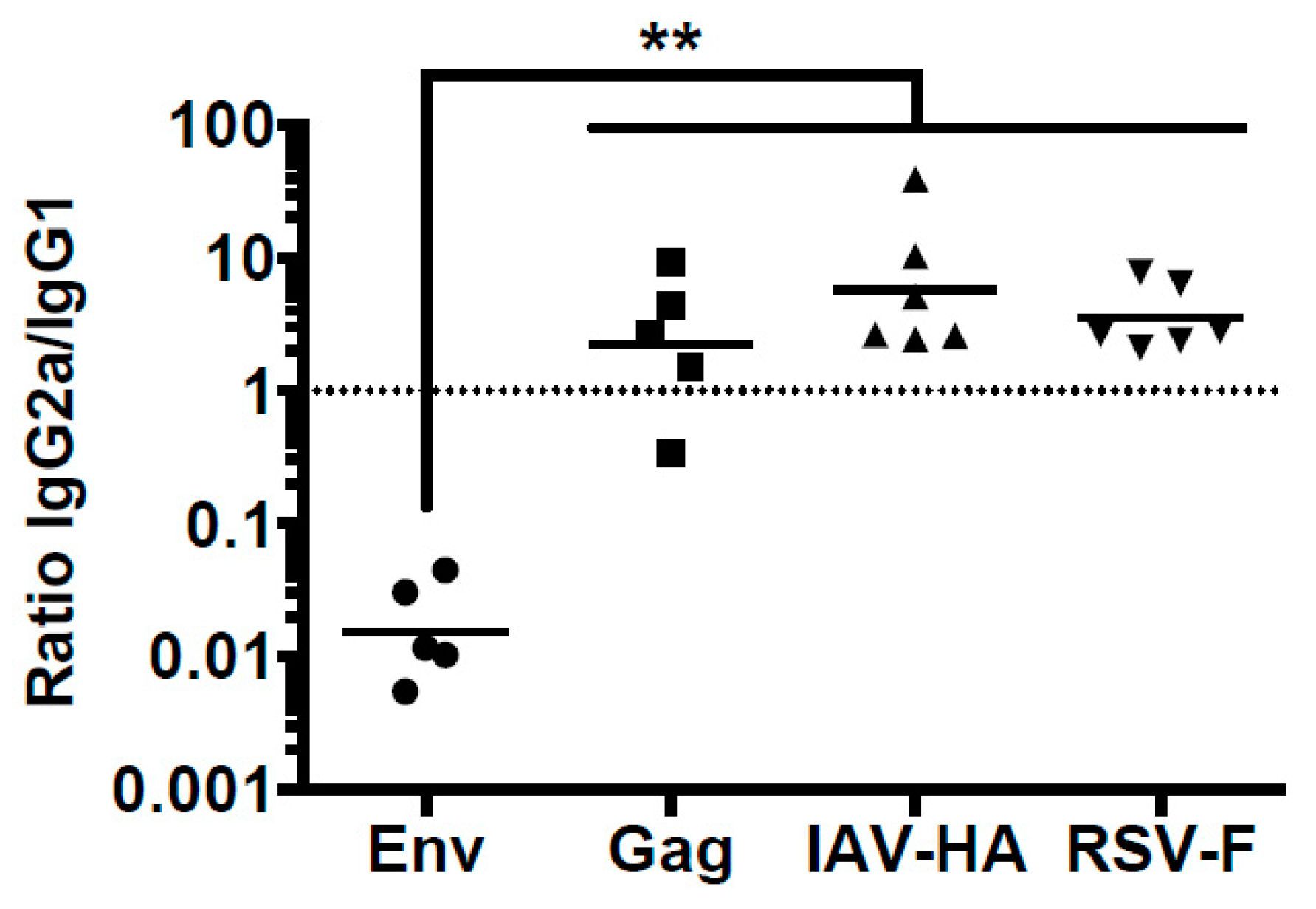
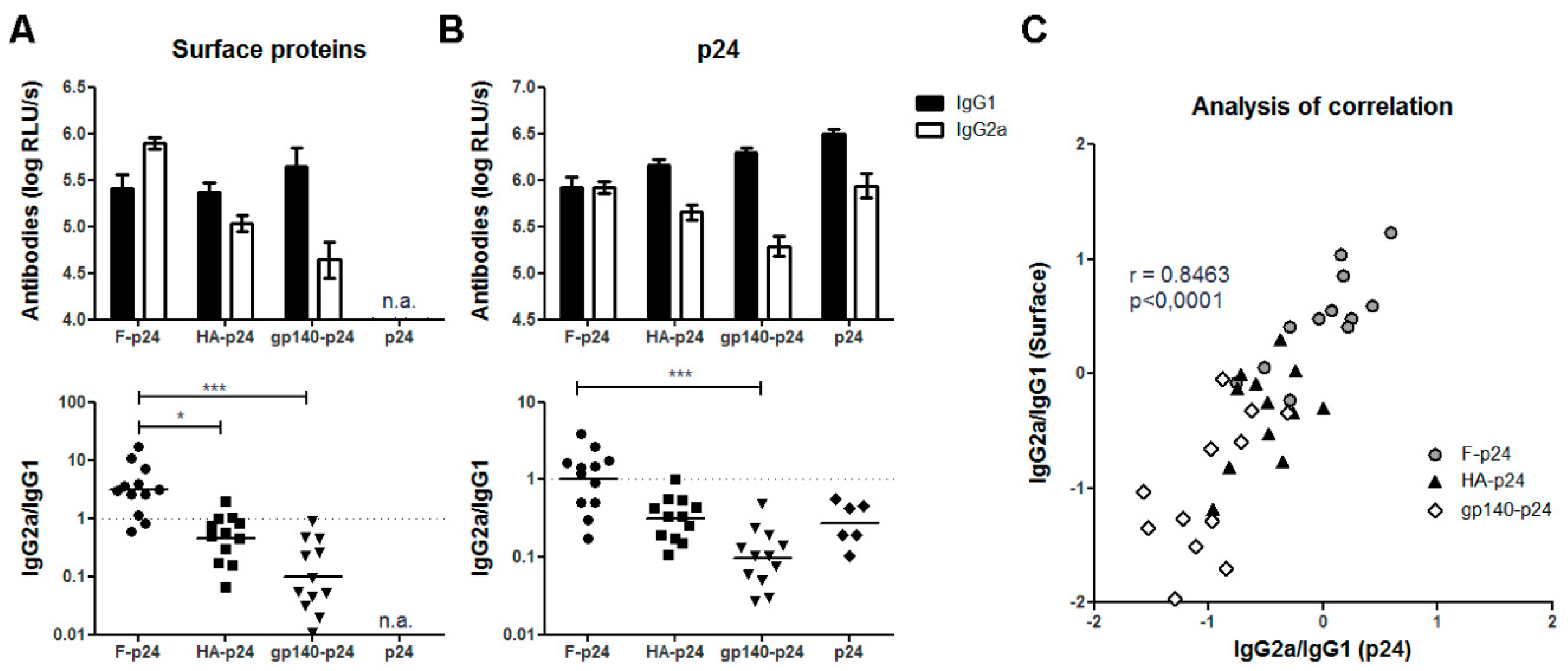
3.1. Differential Antibody Response to Viral Glycoprotein after DNA Immunization
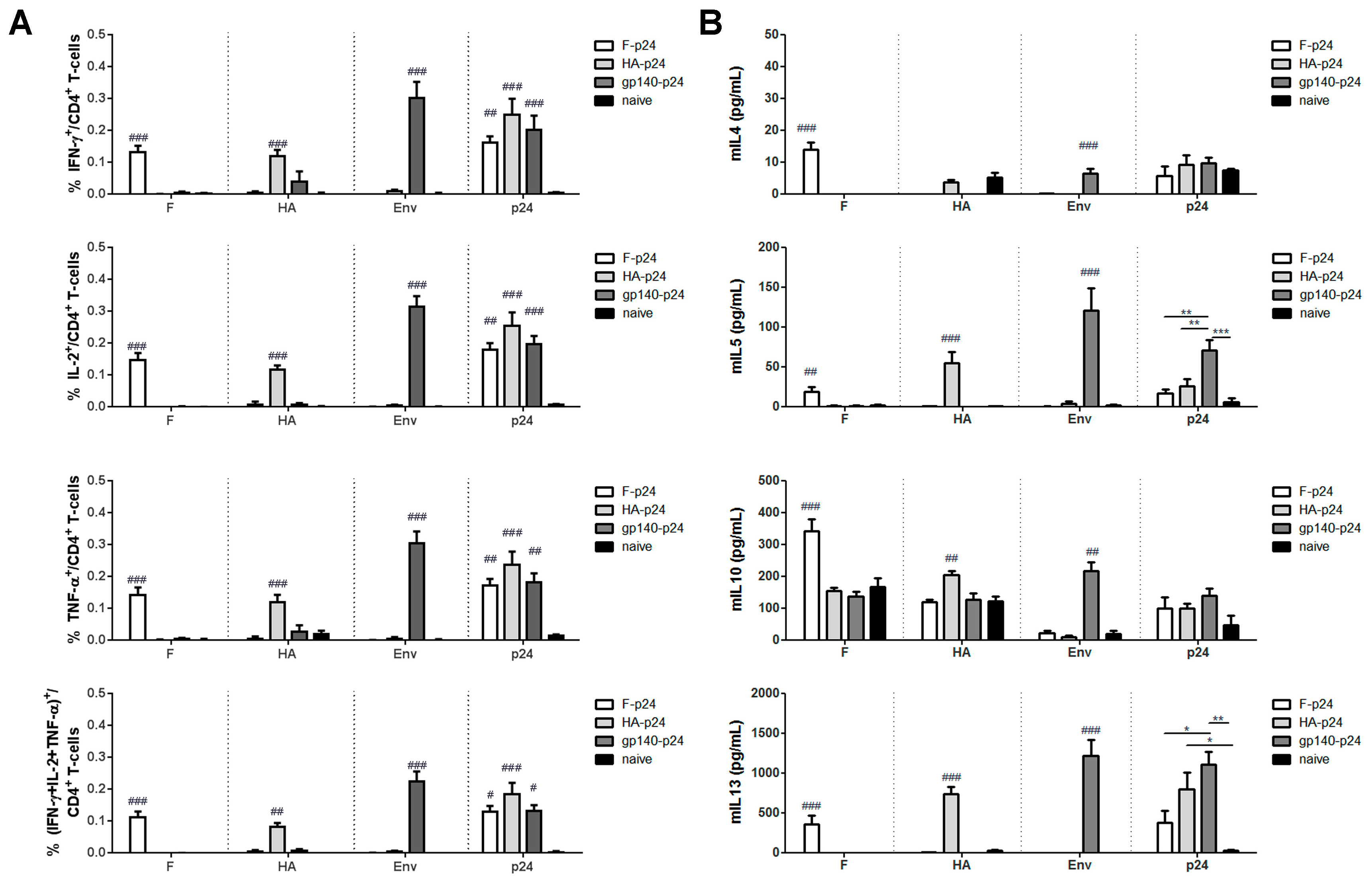
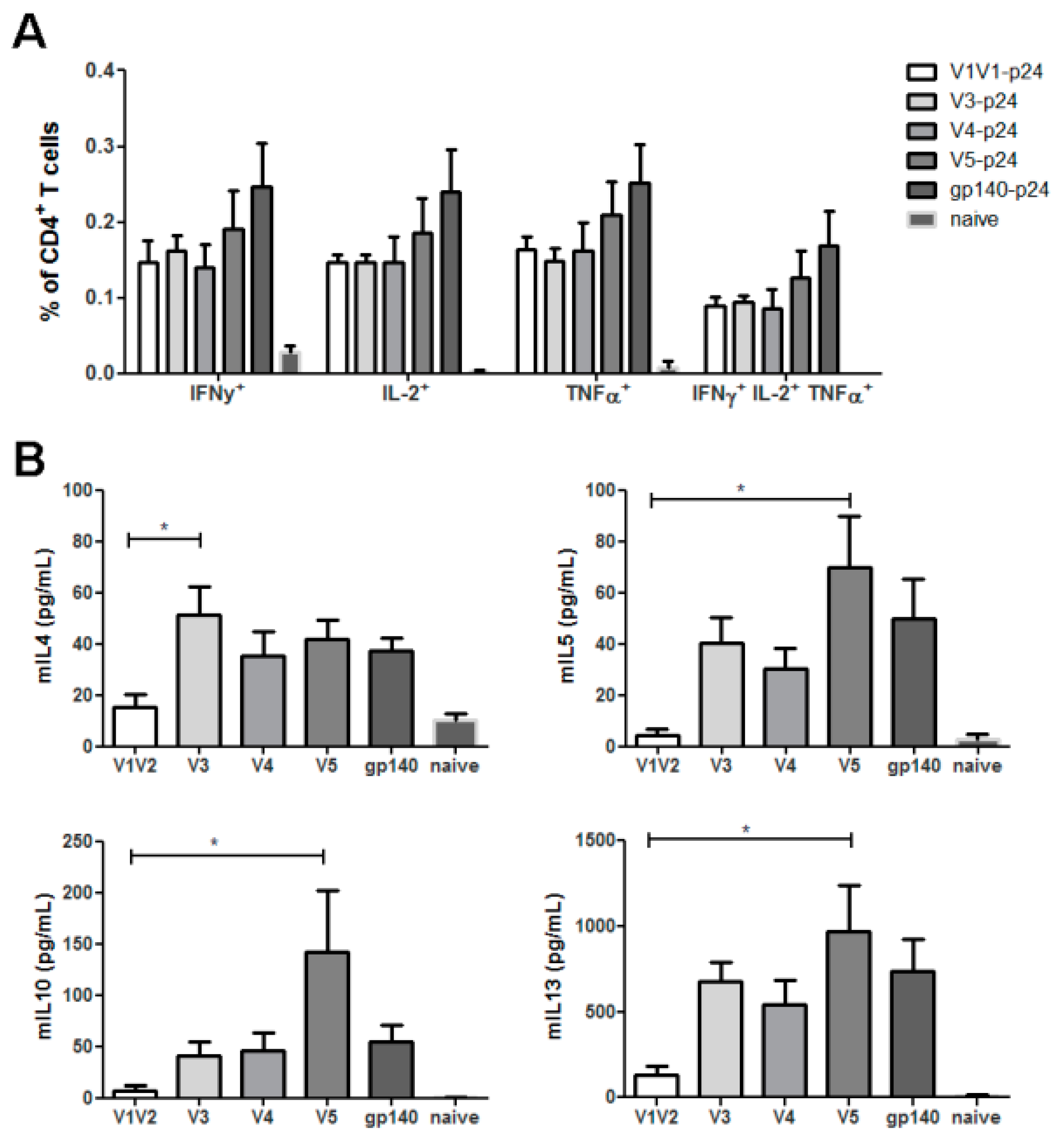
3.2. HIV Env Modulates T-Helper Cell Response
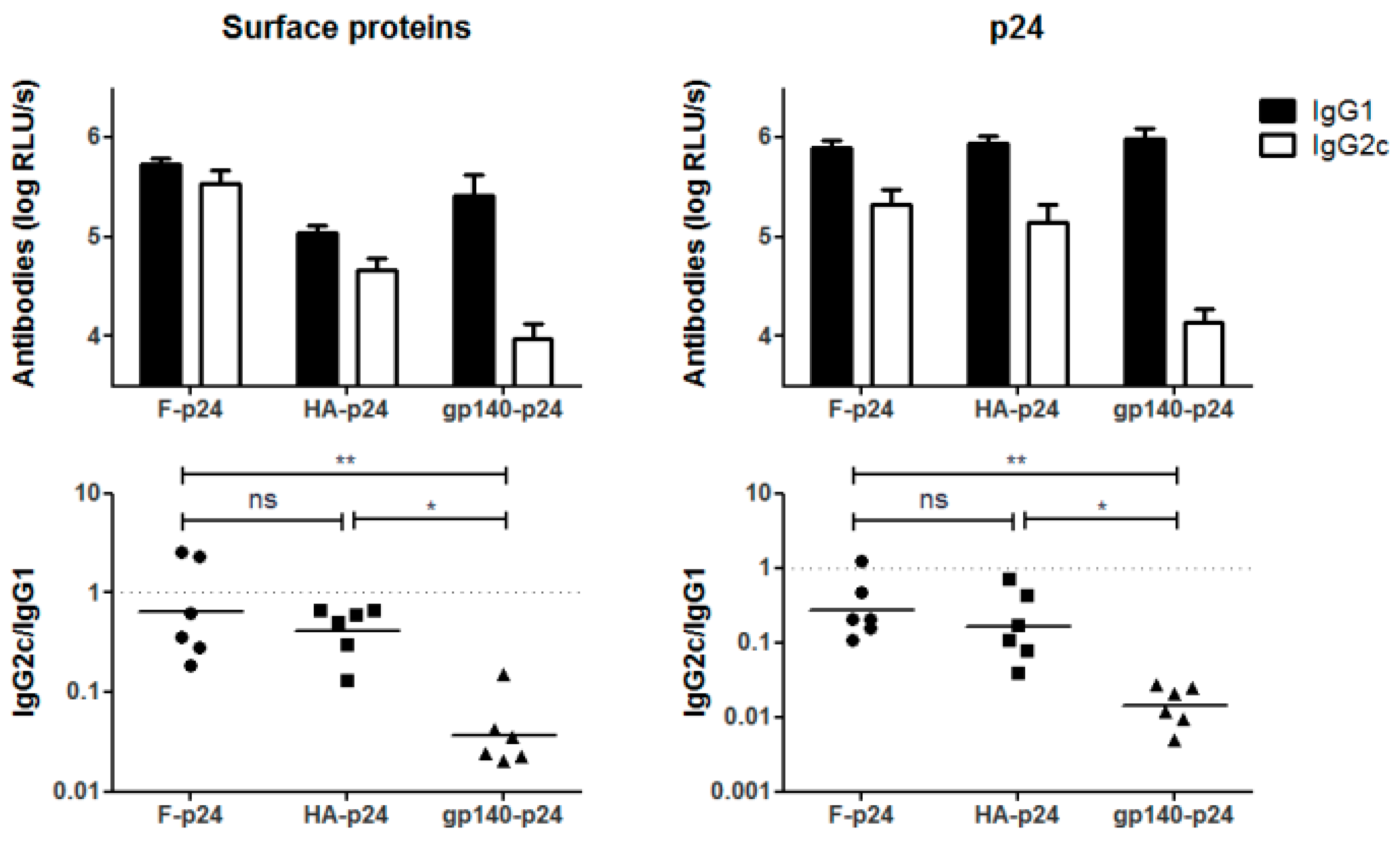
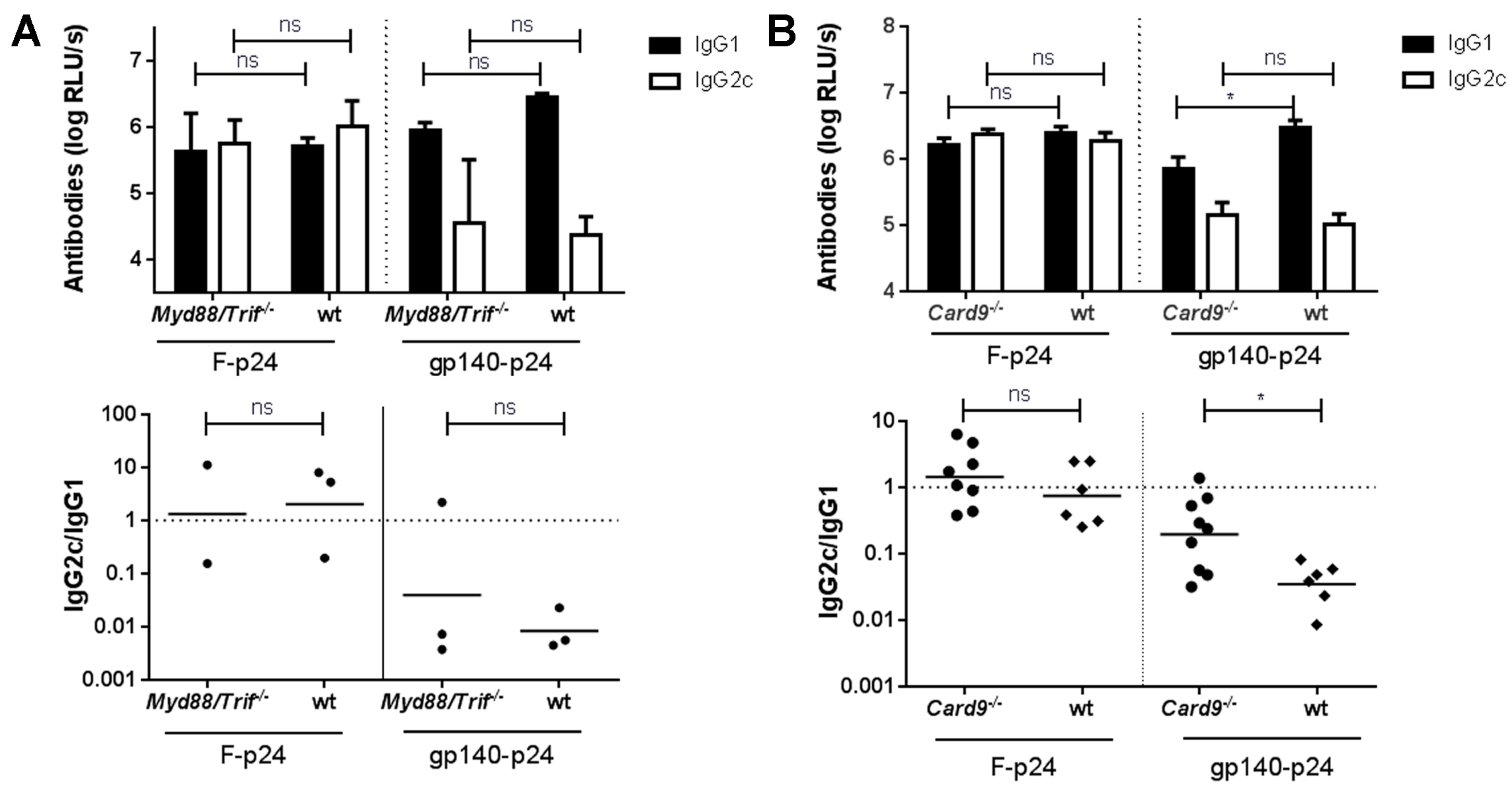
3.3. CLR- but Not TLR-Mediated Signaling Might Be Involved in Env-Mediated Immune Modulation
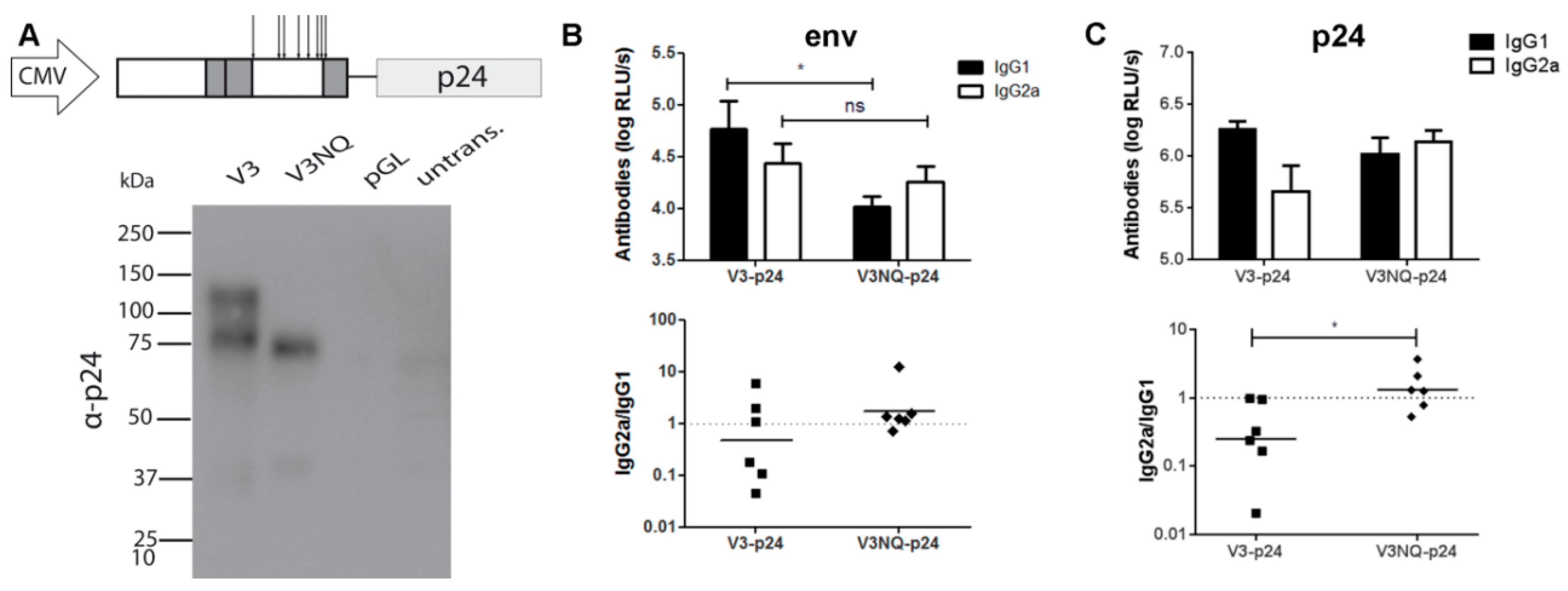
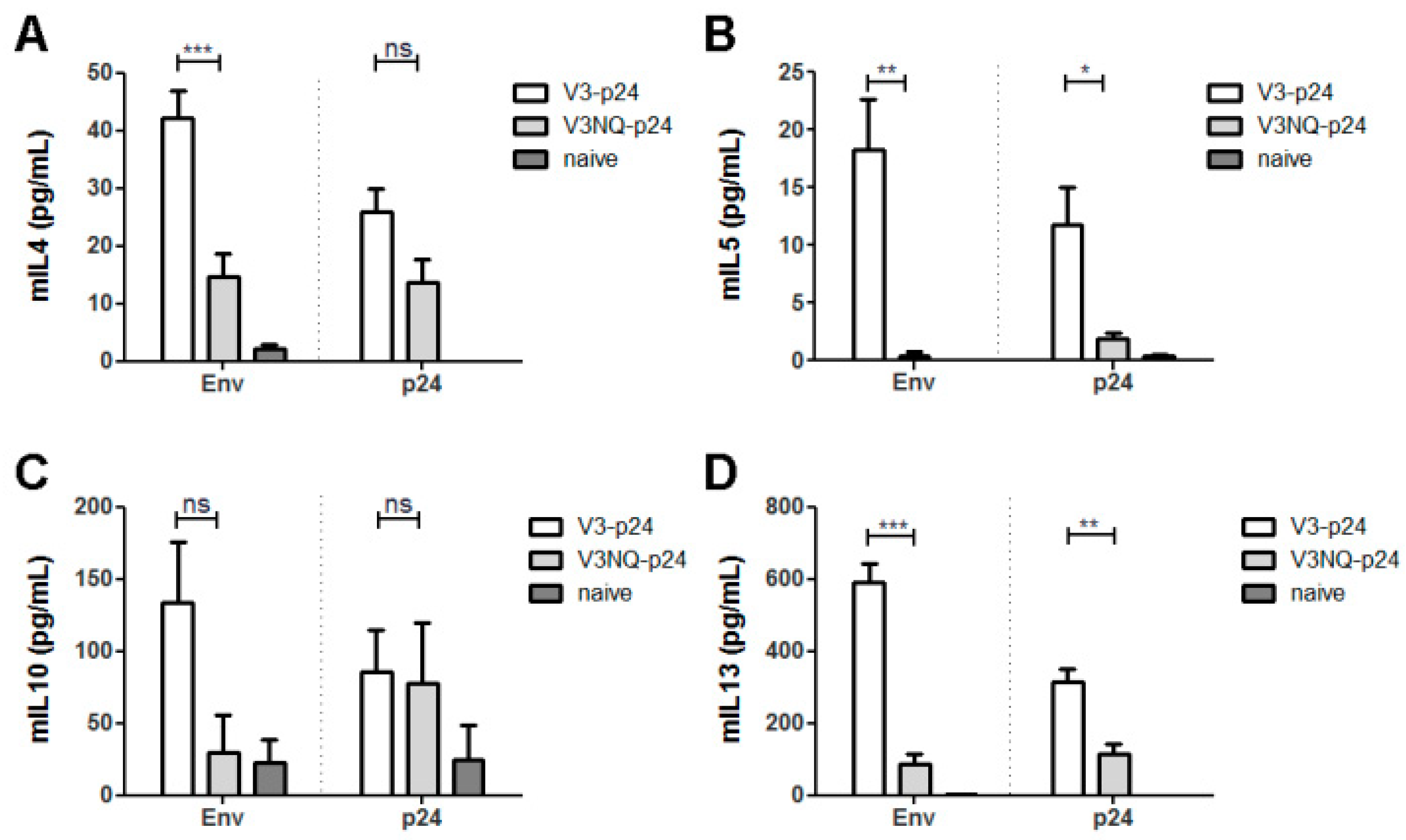
3.4. Glycosylations in the C2V3 Region Determine the IgG1 Bias
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Plotkin, S.A. Vaccines: Correlates of Vaccine-Induced Immunity. Clin. Infect. Dis. 2008, 47, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Yates, N.L.; Liao, H.X.; Fong, Y.; DeCamp, A.; Vandergrift, N.A.; Williams, W.T.; Alam, S.M.; Ferrari, G.; Yang, Z.Y.; Seaton, K.E.; et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci. Transl. Med. 2014, 6, 228ra39. [Google Scholar] [CrossRef] [PubMed]
- Flynn, N.; Forthal, D.; Harro, C.; Judson, F.; Mayer, K.; Para, M. rgp120 HIV Vaccine Study Group Placebo-Controlled Phase 3 Trial of a Recombinant Glycoprotein 120 Vaccine to Prevent HIV-1 Infection. J. Infect. Dis. 2005, 191, 654–665. [Google Scholar] [PubMed]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Immunology: Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 2005, 310, 1510–1512. [Google Scholar] [CrossRef] [PubMed]
- Ljunggren, K.; Broliden, P.A.; Morfeldt-Manson, L.; Jondal, M.; Wahren, B. IgG subclass response to HIV in relation to antibody-dependent cellular cytotoxicity at different clinical stages. Clin. Exp. Immunol. 1988, 73, 343–347. [Google Scholar]
- Bruhns, P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood 2012, 119, 5640–5649. [Google Scholar] [CrossRef] [Green Version]
- Nimmerjahn, F.; Bruhns, P.; Horiuchi, K.; Ravetch, J.V. FcγRIV: A novel FcR with distinct IgG subclass specificity. Immunity 2005, 23, 41–51. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Gordan, S.; Lux, A. FcγR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 2015, 36, 325–336. [Google Scholar] [Green Version]
- Ishizaka, S.T.; Piacente, P.; Silva, J.; Mishkin, E.M. IgG subtype is correlated with efficiency of passive protection and effector function of anti-herpes simplex virus glycoprotein d monoclonal antibodies. J. Infect. Dis. 1995, 172, 1108–1111. [Google Scholar] [CrossRef] [PubMed]
- Markine-Goriaynoff, D.; Coutelier, J.-P. Increased efficacy of the immunoglobulin G2a subclass in antibody-mediated protection against lactate dehydrogenase-elevating virus-induced polioencephalomyelitis revealed with switch mutants. J. Virol. 2002, 76, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Storcksdieck genannt Bonsmann, M.; Niezold, T.; Temchura, V.; Pissani, F.; Ehrhardt, K.; Brown, E.P.; Osei-Owusu, N.Y.; Hannaman, D.; Hengel, H.; Ackerman, M.E.; et al. Enhancing the Quality of Antibodies to HIV-1 Envelope by GagPol-Specific Th Cells. J. Immunol. 2015, 195, 4861–4872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, K.; Andjelic, S.; Klasse, P.J.; Kang, Y.; Sanders, R.W.; Michael, E.; Durso, R.J.; Ketas, T.J.; Olson, W.C.; Moore, J.P. Enzymatic removal of mannose moieties can increase the immune response to HIV-1 gp120 in vivo. Virology 2009, 389, 108–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, L.M.; Johnson, P.A.; Donnelly, G.; Nicolson, C.; Robertson, J.; Mills, K.H.G. Innate IL-10 promotes the induction of Th2 responses with plasmid DNA expressing HIV gp120. Vaccine 2005, 23, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, D.; Caspar, P.; Zweig, M.; Garcia-Moll, M.; Showalter, S.D.; Vogel, F.R.; Sher, A. Adsorption to aluminum hydroxide promotes the activity of IL-12 as an adjuvant for antibody as well as type 1 cytokine responses to HIV-1 gp120. J. Immunol. 1997, 159, 2409–2417. [Google Scholar] [PubMed]
- Moser, M.; Murphy, K.M. Dendritic cell regulation of TH1-TH2 development. Nat. Immunol. 2000, 1, 199–205. [Google Scholar] [CrossRef]
- Murray, J.S. How the MHC selects Th1/Th2 immunity. Immunol. Today 1998, 19, 157–163. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 2016, 16, 433–448. [Google Scholar] [CrossRef]
- Mayer, S.; Raulf, M.K.; Lepenies, B. C-type lectins: Their network and roles in pathogen recognition and immunity. Histochem. Cell Biol. 2017, 147, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Saijo, S.; Ikeda, S.; Yamabe, K.; Kakuta, S.; Ishigame, H.; Akitsu, A.; Fujikado, N.; Kusaka, T.; Kubo, S.; Chung, S.-H.; et al. Dectin-2 recognition of α-mannans and induction of Th17 cell differentiation is essential for host defense against candida albicans. Immunity 2010, 32, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Barrett, N.A.; Rahman, O.M.; Fernandez, J.M.; Parsons, M.W.; Xing, W.; Austen, K.F.; Kanaoka, Y. Dectin-2 mediates Th2 immunity through the generation of cysteinyl leukotrienes. J. Exp. Med. 2011, 208, 593–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gringhuis, S.I.; den Dunnen, J.; Litjens, M.; van der Vlist, M.; Wevers, B.; Bruijns, S.C.M.; Geijtenbeek, T.B.H. Dectin-1 directs T helper cell differentiation by controlling noncannonical NK-kB activation through Raf-1 and Syk. Nat. Immunol. 2009, 10, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Johannssen, T.; Lepenies, B. Glycan-Based Cell Targeting To Modulate Immune Responses. Trends Biotechnol. 2017, 35, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Taoufik, Y.; Lantz, O.; Wallon, C.; Charles, A.; Dussaix, E.; Delfraissy, J.F. Human immunodeficiency virus gp120 inhibits interleukin-12 secretion by human monocytes: An indirect interleukin-10-mediated effect. Blood 1997, 89, 2842–2848. [Google Scholar] [PubMed]
- Cameron, P.U.; Freudenthal, P.S.; Barker, J.M.; Gezelter, S.; Inaba, K.; Steinman, R.M. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 1992, 257, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Shan, M.; Klasse, P.J.; Banerjee, K.; Dey, A.K.; Iyer, S.P.N.; Dionisio, R.; Charles, D.; Campbell-Gardener, L.; Olson, W.C.; Sanders, R.W.; et al. HIV-1 gp120 Mannoses Induce Immunosuppressive Responses from Dendritic Cells. PLoS Pathog. 2007, 3, e169. [Google Scholar] [CrossRef] [PubMed]
- Gringhuis, S.I.; den Dunnen, J.; Litjens, M.; van der Vlist, M.; Geijtenbeek, T.B.H. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 2009, 10, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Graf, M.; Bieler, K.; Wolf, H.; Grunwald, T.; Foley, P.; Uberla, K. Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: Implications for the safety of lentiviral vectors. Hum. Gene Ther. 2000, 11, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Nabi, G.; Genannt Bonsmann, M.; Tenbusch, M.; Gardt, O.; Barouch, D.H.; Temchura, V.; Überla, K. GagPol-specific CD4+ T-cells increase the antibody response to Env by intrastructural help. Retrovirology 2013, 10, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stab, V.; Nitsche, S.; Niezold, T.; Storcksdieck genannt Bonsmann, M.; Wiechers, A.; Tippler, B.; Hannaman, D.; Ehrhardt, C.; Überla, K.; Grunwald, T.; et al. Protective Efficacy and Immunogenicity of a Combinatory DNA Vaccine against Influenza A Virus and the Respiratory Syncytial Virus. PLoS ONE 2013, 8, e72217. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, C.; Tenbusch, M.; Nchinda, G.; Temchura, V.; Nabi, G.; Stone, G.W.; Kornbluth, R.S.; Überla, K. Enhancement of the priming efficacy of DNA vaccines encoding dendritic cell-targeted antigens by synergistic toll-like receptor ligands. BMC Immunol. 2009, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Sanjo, H.; Takeuchi, O.; Sugiyama, M.; Okabe, M.; Takeda, K.; et al. Role of Adaptor TRIF in the MyD88-lndependent Toll-Like Receptor Signaling Pathway. Science 2003, 301, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Gross, O.; Gewies, A.; Finger, K.; Schäfer, M.; Sparwasser, T.; Peschel, C.; Förster, I.; Ruland, J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 2006, 442, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.W.; Ghebremichael, M.; Robinson, H.; Brown, E.; Choi, I.; Lane, S.; Dugast, A.-S.; Schoen, M.K.; Rolland, M.; Suscovich, T.J.; et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci. Transl. Med. 2014, 6, 228ra38. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, K.; Klasse, P.J.; Sanders, R.W.; Pereyra, F.; Michael, E.; Lu, M.; Walker, B.D.; Moore, J.P. IgG subclass profiles in infected HIV type 1 controllers and chronic progressors and in uninfected recipients of Env vaccines. AIDS Res. Hum. Retroviruses 2010, 26, 445–458. [Google Scholar] [CrossRef]
- Nazli, A.; Kafka, J.K.; Ferreira, V.H.; Anipindi, V.; Mueller, K.; Osborne, B.J.; Dizzell, S.; Chauvin, S.; Mian, M.F.; Ouellet, M.; et al. HIV-1 gp120 Induces TLR2- and TLR4-Mediated Innate Immune Activation in Human Female Genital Epithelium. J. Immunol. 2013, 191, 4246–4258. [Google Scholar] [CrossRef] [Green Version]
- Haynes, L.M.; Moore, D.D.; Kurt-Jones, E.A.; Finberg, R.W.; Anderson, L.J.; Tripp, R.A. Involvement of toll-like receptor 4 in innate immunity to respiratory syncytial virus. J. Virol. 2001, 75, 10730–10737. [Google Scholar] [CrossRef]
- Decout, A.; Silva-Gomes, S.; Drocourt, D.; Barbe, S.; André, I.; Cueto, F.J.; Lioux, T.; Sancho, D.; Pérouzel, E.; Vercellone, A.; et al. Rational design of adjuvants targeting the C-type lectin Mincle. Proc. Natl. Acad. Sci. USA 2017, 114, 2675–2680. [Google Scholar] [CrossRef]
- Zou, Z.; Chastain, A.; Moir, S.; Ford, J.; Trandem, K.; Martinelli, E.; Cicala, C.; Crocker, P.; Arthos, J.; Sun, P.D. Siglecs facilitate HIV-1 infection of macrophages through adhesion with viral sialic acids. PLoS ONE 2011, 6, e24559. [Google Scholar] [CrossRef] [PubMed]
- Sewald, X.; Ladinsky, M.S.; Uchil, P.D.; Beloor, J.; Pi, R.; Herrmann, C.; Motamedi, N.; Murooka, T.T.; Brehm, M.A.; Greiner, D.L.; et al. Retroviruses use CD169-mediated trans-infection of permissive lymphocytes to establish infection. Science 2015, 350, 563–567. [Google Scholar] [CrossRef]
- Chang, Y.C.; Olson, J.; Beasley, F.C.; Tung, C.; Zhang, J.; Crocker, P.R.; Varki, A.; Nizet, V. Group B Streptococcus Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses In Vivo. PLoS Pathog. 2014, 10, e1003846. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.F.; Uchiyama, S.; Chang, Y.C.; Lewis, A.L.; Nizet, V.; Varki, A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 2009, 113, 3333–3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggink, D.; Melchers, M.; Wuhrer, M.; van Montfort, T.; Dey, A.K.; Naaijkens, B.A.; David, K.B.; Le Douce, V.; Deelder, A.M.; Kang, K.; et al. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology 2010, 401, 236–247. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Lin, Y.; Spearman, M.; Cheng, P.; Butler, M.; Wu, S. Influenza Virus Hemagglutinin Glycoproteins with Different N-Glycan Patterns Activate Dendritic Cells In Vitro. J. Virol. 2016, 90, 6085–6096. [Google Scholar] [CrossRef]
- Gupta, S.; Clark, E.S.; Termini, J.M.; Boucher, J.; Kanagavelu, S.; LeBranche, C.C.; Abraham, S.; Montefiori, D.C.; Khan, W.N.; Stone, G.W. DNA Vaccine Molecular Adjuvants SP-D-BAFF and SP-D-APRIL Enhance Anti-gp120 Immune Response and Increase HIV-1 Neutralizing Antibody Titers. J. Virol. 2015, 89, 4158–4169. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Qiao, X.; Klasse, P.J.; Chiu, A.; Chadburn, A.; Knowles, D.M.; Moore, J.P.; Cerutti, A. HIV-1 Envelope Triggers Polyclonal Ig Class Switch Recombination through a CD40-Independent Mechanism Involving BAFF and C-Type Lectin Receptors. J. Immunol. 2006, 176, 3931–3941. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heß, R.; Storcksdieck genannt Bonsmann, M.; Lapuente, D.; Maaske, A.; Kirschning, C.; Ruland, J.; Lepenies, B.; Hannaman, D.; Tenbusch, M.; Überla, K. Glycosylation of HIV Env Impacts IgG Subtype Responses to Vaccination. Viruses 2019, 11, 153. https://doi.org/10.3390/v11020153
Heß R, Storcksdieck genannt Bonsmann M, Lapuente D, Maaske A, Kirschning C, Ruland J, Lepenies B, Hannaman D, Tenbusch M, Überla K. Glycosylation of HIV Env Impacts IgG Subtype Responses to Vaccination. Viruses. 2019; 11(2):153. https://doi.org/10.3390/v11020153
Chicago/Turabian StyleHeß, Rebecca, Michael Storcksdieck genannt Bonsmann, Dennis Lapuente, Andre Maaske, Carsten Kirschning, Jürgen Ruland, Bernd Lepenies, Drew Hannaman, Matthias Tenbusch, and Klaus Überla. 2019. "Glycosylation of HIV Env Impacts IgG Subtype Responses to Vaccination" Viruses 11, no. 2: 153. https://doi.org/10.3390/v11020153




