Cytotoxicity and Transcriptomic Analyses of Biogenic Palladium Nanoparticles in Human Ovarian Cancer Cells (SKOV3)
Abstract
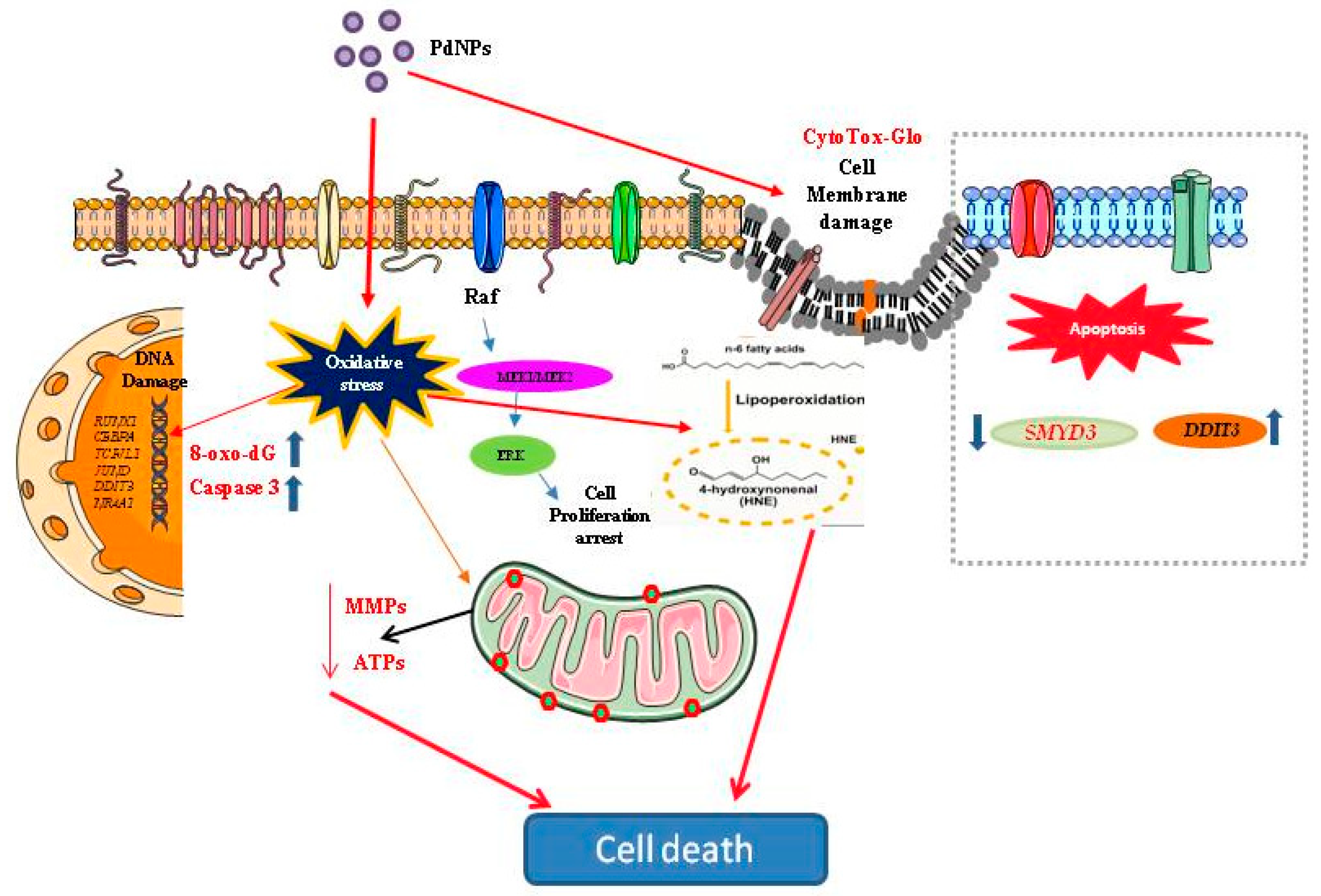
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Characterization of PdNPs
2.2. Cell Viability and Cell Proliferation
2.3. Membrane Integrity
2.4. Assessment of Dead-Cell Protease Activity
2.5. Determination of Intracellular ROS
2.6. Measurement of MDA Content and Antioxidant Enzyme Activities
2.7. JC-1 Assay
2.8. Measurement of ATP Levels
2.9. Measurement of 8-oxo-dG
2.10. Measurement of Caspase 3 Activity
2.11. Cell Cycle Analysis
2.12. RNA-Seq and Bioinformatics Analysis
2.13. Pathway Analysis
2.14. Statistical Methods
3. Results and Discussion
3.1. Synthesis and Characterization of PdNPs
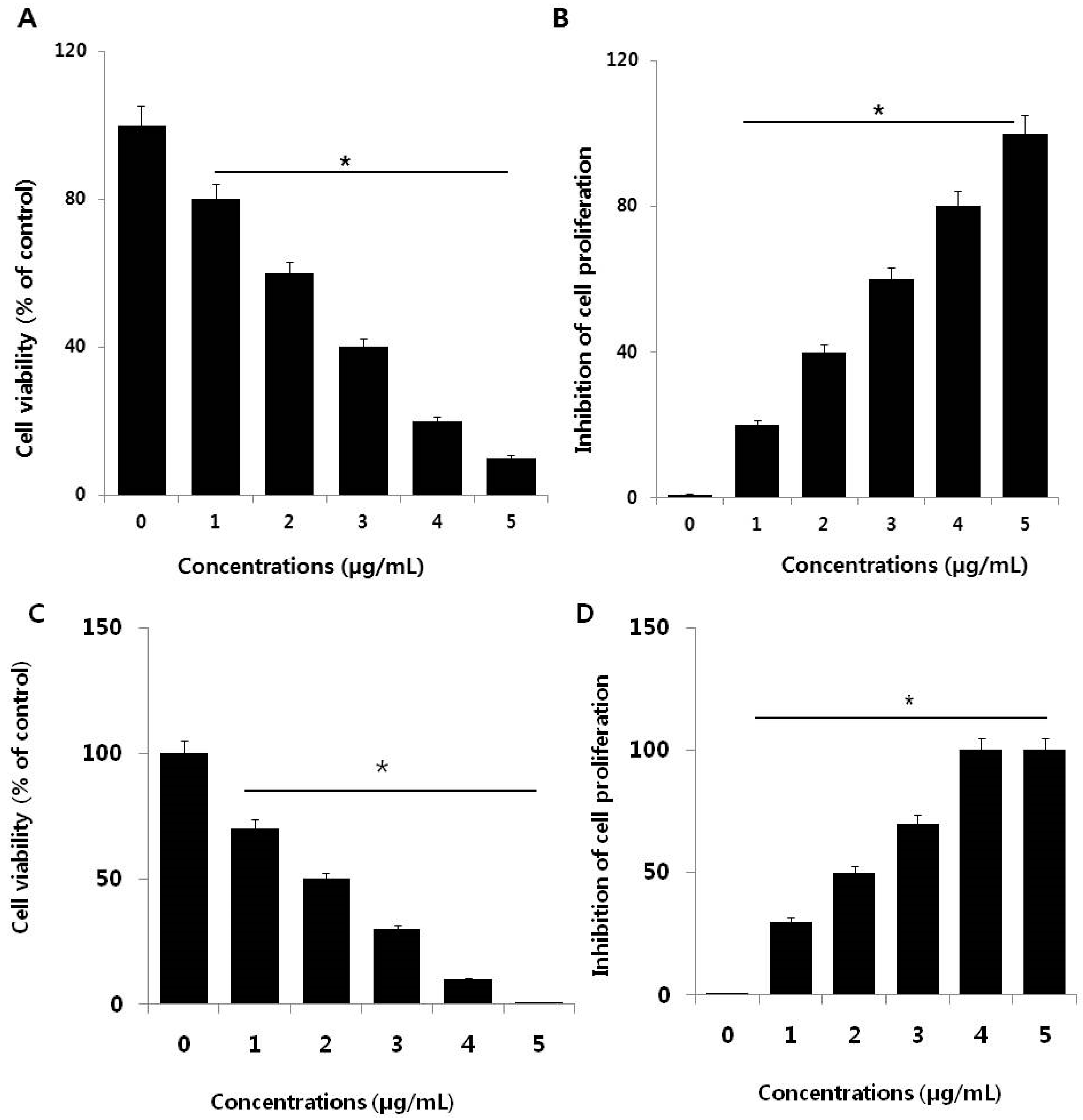
3.2. PdNPs Inhibit Cell Survival and Proliferation
3.3. PdNPs Induce Leakage of LDH and Decrease Dead-Cell Protease Activity in SKOV3 Cells
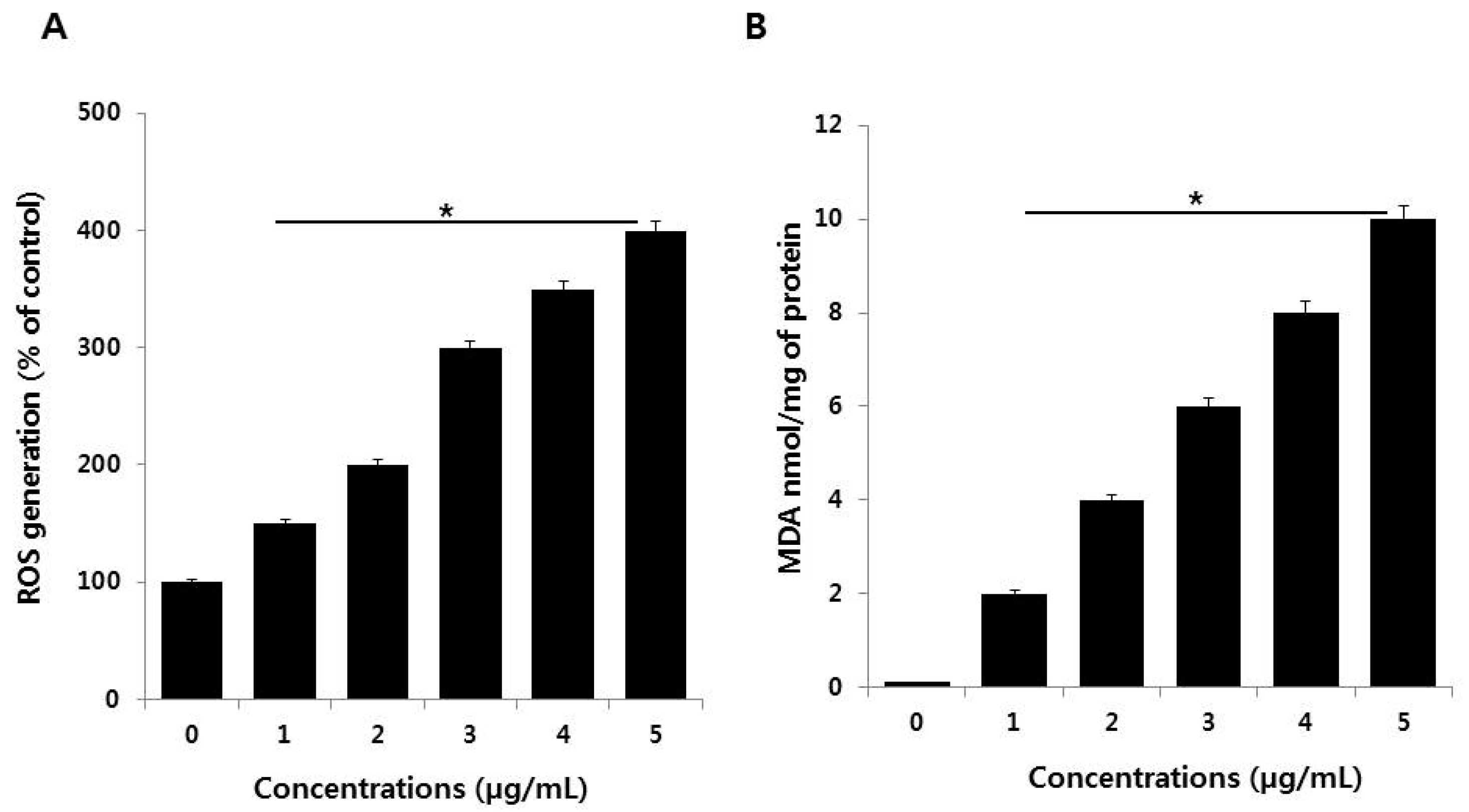
3.4. PdNPs Induce Oxidative Stress and Increase MDA Levels
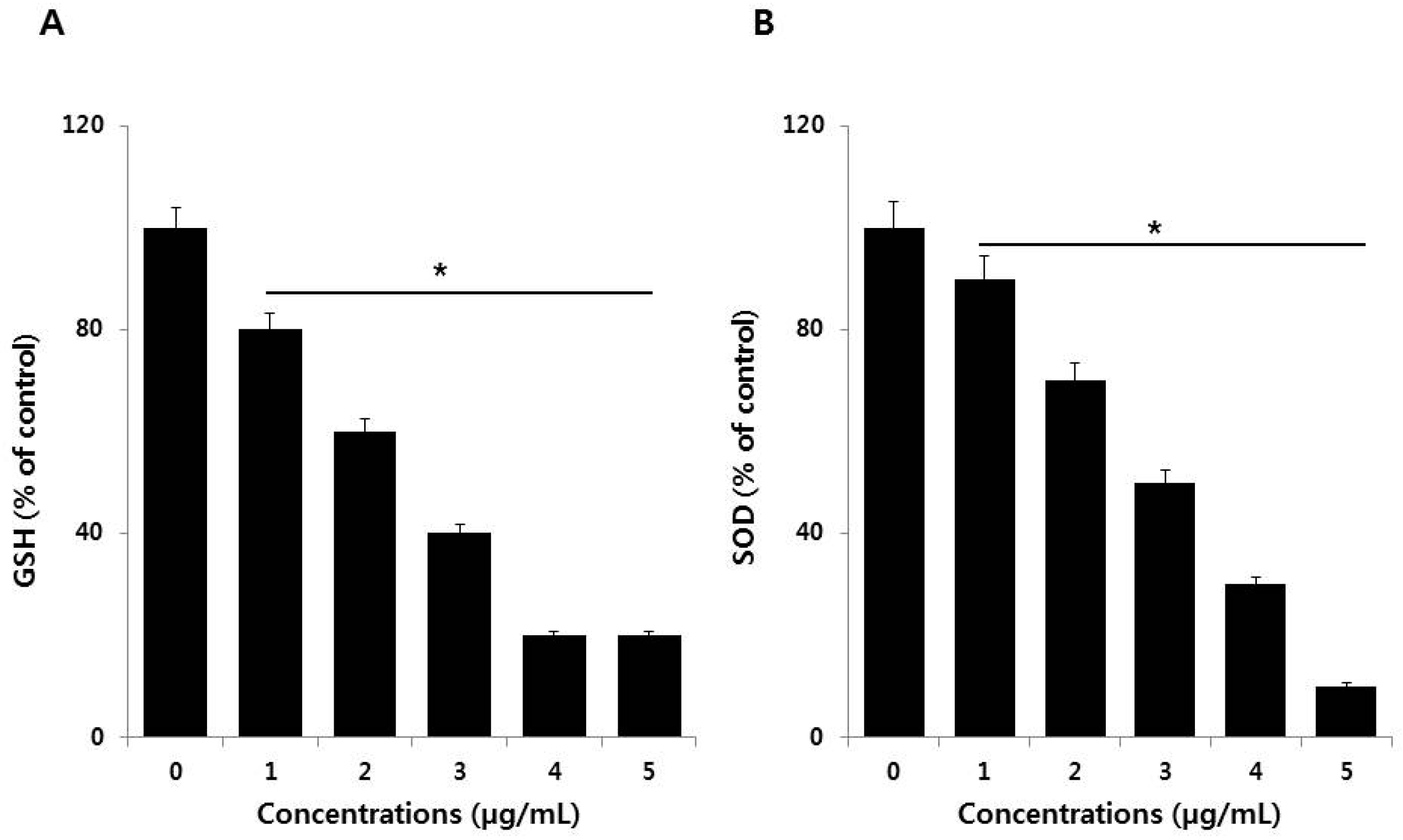
3.5. Effect of PdNPs on Antioxidants
3.6. PdNPs Induce Mitochondrial Dysfunction and Reduce ATP Generation
3.7. PdNPs Induce DNA Damage and Caspase 3 Activation
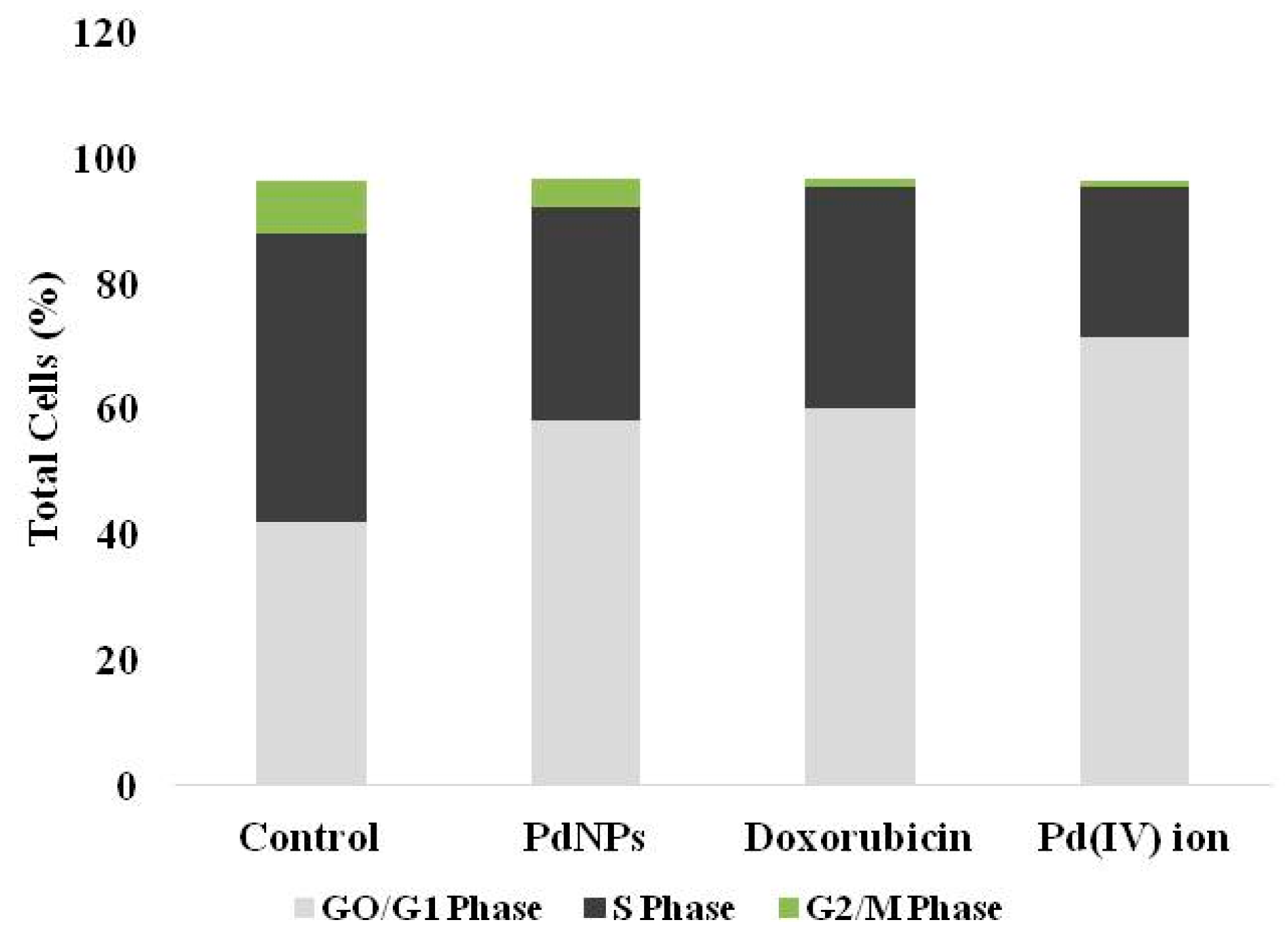
3.8. Effect of PdNPs on Cell Cycle
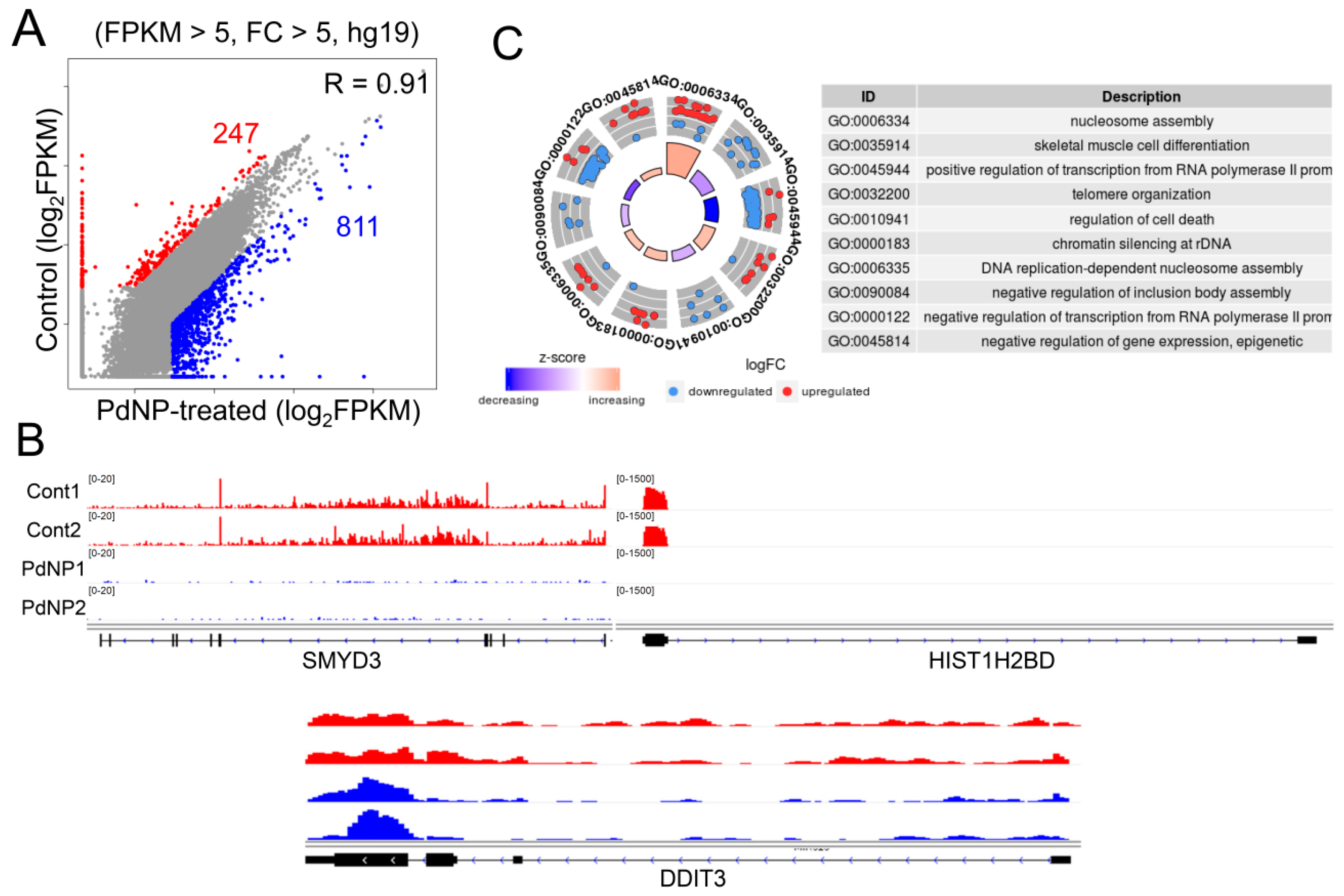
3.9. PdNPs Exposure Alters the Expression Patterns of Multiple Genes in SKOV3 Cells
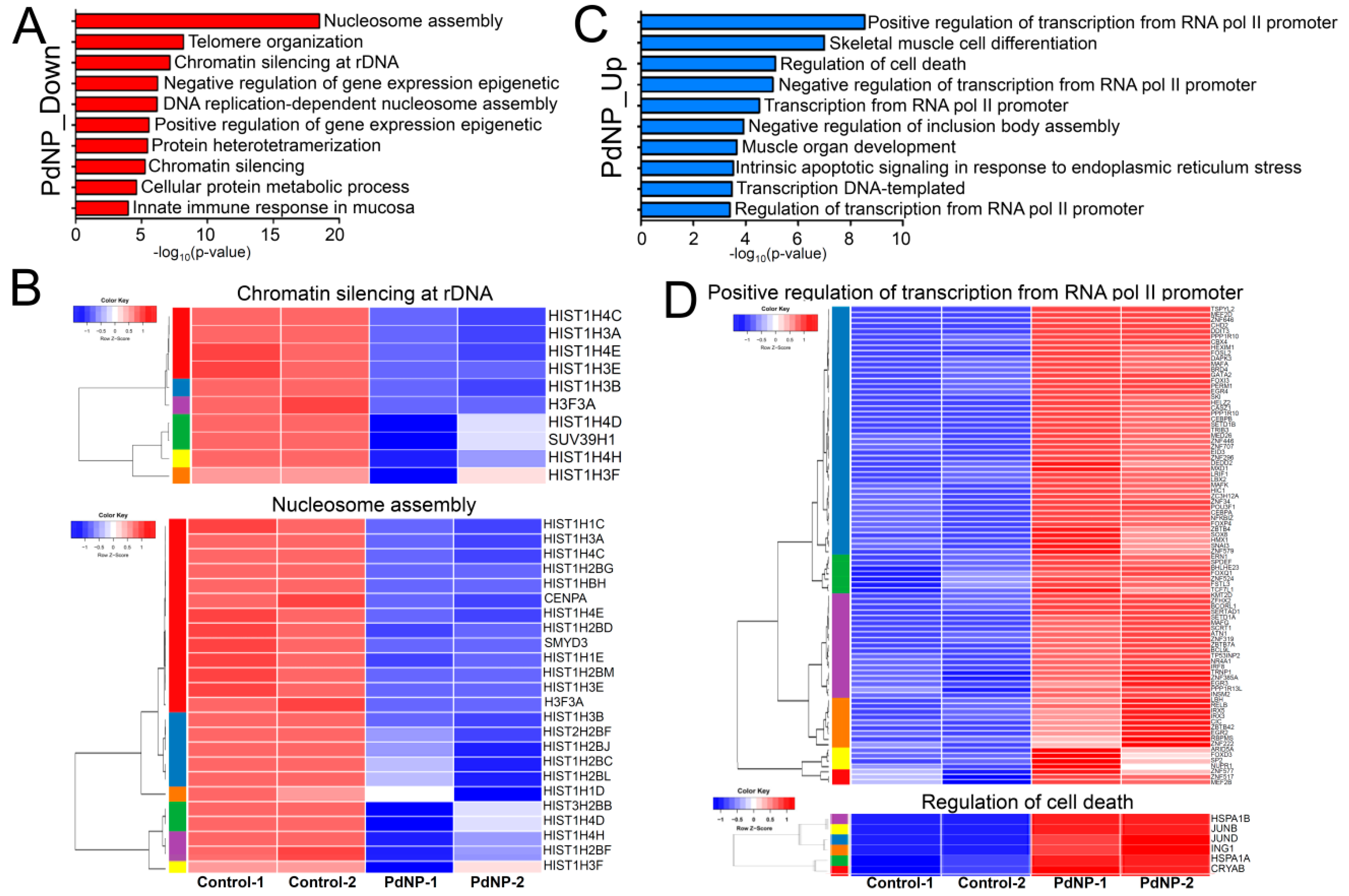
3.10. Genes Involved in Epigenetic Regulation and Apoptosis Were Aberrantly Expressed after PdNPs Exposure
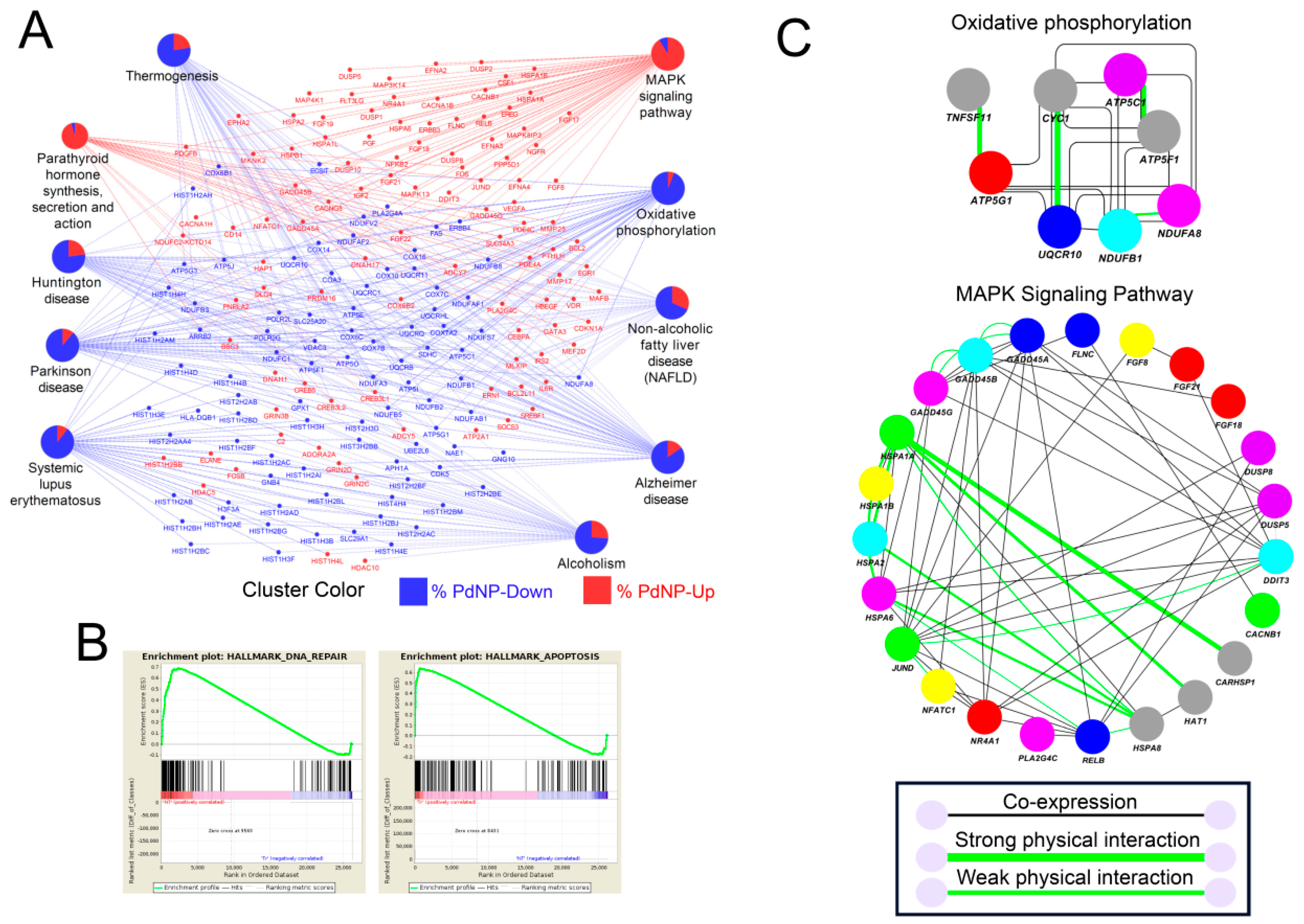
3.11. Treatment with PdNPs-Altered Biological Pathways Involved in Human Disease and Apoptosis
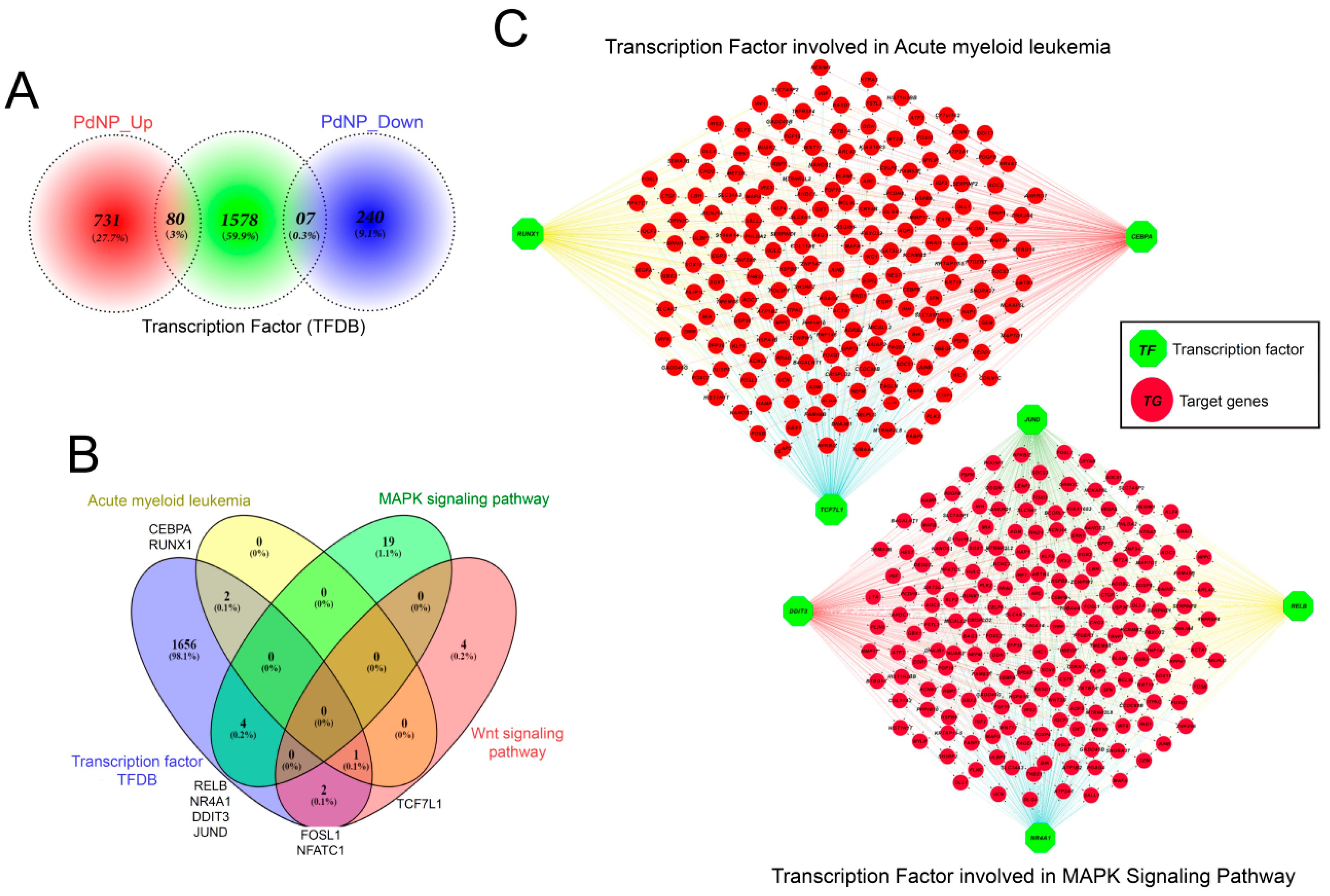
3.12. Transcription Factors Associated with Apoptosis Were Aberrantly Enhanced by PdNPs
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nguyen, P.L.; Gu, X.; Lipsitz, S.R.; Choueiri, T.K.; Choi, W.W.; Lei, Y.; Hoffman, K.E.; Hu, J.C. Cost Implications of the Rapid Adoption of Newer Technologies for Treating Prostate Cancer. J. Clin. Oncol. 2011, 29, 1517–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignani, S.; Bryszewska, M.; Klajnert-Maculewicz, B.; Zablocka, M.; Majoral, J.-P. Advances in Combination Therapies Based on Nanoparticles for Efficacious Cancer Treatment: An Analytical Report. Biomacromolecules 2015, 16, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, R.; Roseblade, A.; Hansbro, P.M.; Rathbone, M.J.; Dua, K.; Bebawy, M. Nanoparticles in Cancer Treatment: Opportunities and Obstacles. Curr. Drug Targets 2018, 19, 1696–1709. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Huang, F.-H.; Zhang, G.-L.; Bai, D.-P.; Massimo, D.F.; Huang, Y.-F.; Gurunathan, S. Novel biomolecule lycopene-reduced graphene oxide-silver nanoparticle enhances apoptotic potential of trichostatin A in human ovarian cancer cells (SKOV3). Int. J. Nanomedicine 2017, 12, 7551–7575. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.; Park, J.H.; Kim, J.-H. A green chemistry approach for synthesizing biocompatible gold nanoparticles. Nanoscale Res. Lett. 2014, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kalishwaralal, K.; Vaidyanathan, R.; Venkataraman, D.; Pandian, S.R.K.; Muniyandi, J.; Hariharan, N.; Eom, S.H. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf. B Biointerfaces 2009, 74, 328–335. [Google Scholar] [CrossRef]
- Han, J.W.; Jeong, J.-K.; Gurunathan, S.; Choi, Y.-J.; Das, J.; Kwon, D.-N.; Cho, S.-G.; Park, C.; Seo, H.G.; Park, J.-K.; et al. Male- and female-derived somatic and germ cell-specific toxicity of silver nanoparticles in mouse. Nanotoxicology 2016, 10, 361–373. [Google Scholar] [CrossRef]
- Das, P.; Linert, W. Schiff base-derived homogeneous and heterogeneous palladium catalysts for the Suzuki–Miyaura reaction. Coord. Chem. Rev. 2016, 311, 1–23. [Google Scholar] [CrossRef]
- Davis, S.E.; Ide, M.S.; Davis, R.J. Selective oxidation of alcohols and aldehydes over supported metal nanoparticles. Green Chem. 2013, 15, 17–45. [Google Scholar] [CrossRef]
- Aditya, T.; Pal, A.; Pal, T. Nitroarene reduction: A trusted model reaction to test nanoparticle catalysts. Chem. Commun. (Camb). 2015, 51, 9410–9431. [Google Scholar] [CrossRef]
- Leso, V.; Iavicoli, I. Palladium Nanoparticles: Toxicological Effects and Potential Implications for Occupational Risk Assessment. Int. J. Mol. Sci. 2018, 19, 503. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Khan, M.; Kuniyil, M.; Adil, S.F.; Al-Warthan, A.; Alkhathlan, H.Z.; Tremel, W.; Tahir, M.N.; Siddiqui, M.R.H. Biogenic synthesis of palladium nanoparticles using Pulicaria glutinosa extract and their catalytic activity towards the Suzuki coupling reaction. Dalton Trans. 2014, 43, 9026–9031. [Google Scholar] [CrossRef]
- Basavegowda, N.; Mishra, K.; Lee, Y.R. Ultrasonic-assisted green synthesis of palladium nanoparticles and their nanocatalytic application in multicomponent reaction. New J. Chem. 2015, 39, 972–977. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kim, E.; Han, J.; Park, J.; Kim, J.-H. Green Chemistry Approach for Synthesis of Effective Anticancer Palladium Nanoparticles. Molecules 2015, 20, 22476–22498. [Google Scholar] [CrossRef]
- Hazarika, M.; Borah, D.; Bora, P.; Silva, A.R.; Das, P. Biogenic synthesis of palladium nanoparticles and their applications as catalyst and antimicrobial agent. PLoS ONE 2017, 12, e0184936. [Google Scholar] [CrossRef]
- Bankar, A.; Joshi, B.; Kumar, A.R.; Zinjarde, S. Banana peel extract mediated novel route for the synthesis of palladium nanoparticles. Mater. Lett. 2010, 64, 1951–1953. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Mohammad Sajadi, S. Pd nanoparticles synthesized in situ with the use of Euphorbia granulate leaf extract: Catalytic properties of the resulting particles. J. Colloid Interface Sci. 2016, 462, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Saikia, P.K.; Bhattacharjee, R.P.; Sarmah, P.P.; Saikia, L.; Dutta, D.K. A green synthesis of Pd nanoparticles supported on modified montmorillonite using aqueous Ocimum sanctum leaf extract: A sustainable catalyst for hydrodechlorination of 4-chlorophenol. RSC Adv. 2016, 6, 110011–110018. [Google Scholar] [CrossRef]
- Wilkinson, K.E.; Palmberg, L.; Witasp, E.; Kupczyk, M.; Feliu, N.; Gerde, P.; Seisenbaeva, G.A.; Fadeel, B.; Dahlén, S.E.; Kessler, V.G. Solution-engineered palladium nanoparticles: Model for health effect studies of automotive particulate pollution. ACS Nano 2011, 5, 5312–5324. [Google Scholar] [CrossRef] [PubMed]
- Petrarca, C.; Clemente, E.; Di Giampaolo, L.; Mariani-Costantini, R.; Leopold, K.; Schindl, R.; Lotti, L.V.; Mangifesta, R.; Sabbioni, E.; Niu, Q.; et al. Palladium nanoparticles induce disturbances in cell cycle entry and progression of peripheral blood mononuclear cells: Paramount role of ions. J. Immunol. Res. 2014, 2014, 295092. [Google Scholar] [CrossRef]
- Ghosh, S.; Nitnavare, R.; Dewle, A.; Tomar, G.B.; Chippalkatti, R.; More, P.; Kitture, R.; Kale, S.; Bellare, J.; Chopade, B.A. Novel platinum-palladium bimetallic nanoparticles synthesized by Dioscorea bulbifera: Anticancer and antioxidant activities. Int. J. Nanomedicine 2015, 10, 7477–7490. [Google Scholar] [PubMed]
- Pan, Y.-T.; Smith, C.E.; Kwok, K.S.; Chen, J.; Kong, H.; Yang, H. Functionalized ultrathin palladium nanosheets as patches for HepG2 cancer cells. Chem. Commun. (Camb). 2015, 51, 14171–14174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarifi, S.; Ali, D.; Alkahtani, S.; Almeer, R.S. ROS-Mediated Apoptosis and Genotoxicity Induced by Palladium Nanoparticles in Human Skin Malignant Melanoma Cells. Oxid. Med. Cell. Longev. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhusseiny, A.F.; Hassan, H.H.A.M. Antimicrobial and antitumor activity of platinum and palladium complexes of novel spherical aramides nanoparticles containing flexibilizing linkages: Structure–property relationship. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 103, 232–245. [Google Scholar] [CrossRef]
- Boscolo, P.; Bellante, V.; Leopold, K.; Maier, M.; Di Giampaolo, L.; Antonucci, A.; Iavicoli, I.; Tobia, L.; Paoletti, A.; Montalti, M.; et al. Effects of palladium nanoparticles on the cytokine release from peripheral blood mononuclear cells of non-atopic women. J. Biol. Regul. Homeost. Agents 2010, 24, 207–214. [Google Scholar]
- Reale, M.; Vianale, G.; Lotti, L.V.; Mariani-Costantini, R.; Perconti, S.; Cristaudo, A.; Leopold, K.; Antonucci, A.; Di Giampaolo, L.; Iavicoli, I.; et al. Effects of Palladium Nanoparticles on the Cytokine Release from Peripheral Blood Mononuclear Cells of Palladium-Sensitized Women. J. Occup. Environ. Med. 2011, 53, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Leso, V.; Marinaccio, A.; Cenacchi, G.; Papa, V.; Leopold, K.; Schindl, R.; Bocca, B.; Alimonti, A.; Iavicoli, I. The effects of palladium nanoparticles on the renal function of female Wistar rats. Nanotoxicology 2015, 9, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, H.; Li, Y.; Ruan, Y.; Quan, C. Identification of genes and pathways associated with MDR in MCF-7/MDR breast cancer cells by RNA-seq analysis. Mol. Med. Rep. 2018, 17, 6211–6226. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.-X.; Li, B.-L.; Liu, X.-H.; Yuan, Y.; Lu, C.-J.; Chen, R.; Zhao, J. RNA-seq reveals determinants of sensitivity to chemotherapy drugs in esophageal carcinoma cells. Int. J. Clin. Exp. Pathol. 2014, 7, 1524–1533. [Google Scholar]
- Wacker, S.A.; Houghtaling, B.R.; Elemento, O.; Kapoor, T.M. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat. Chem. Biol. 2012, 8, 235–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Silver nanoparticles enhance the apoptotic potential of gemcitabine in human ovarian cancer cells: Combination therapy for effective cancer treatment. Int. J. Nanomedicine 2017, 12, 6487–6502. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Jeyaraj, M.; Kim, J.-H. Cytotoxicity of biologically synthesized silver nanoparticles in MDA-MB-231 human breast cancer cells. Biomed Res. Int. 2013, 2013, 535796. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kim, J.-H. Graphene Oxide–Silver Nanoparticles Nanocomposite Stimulates Differentiation in Human Neuroblastoma Cancer Cells (SH-SY5Y). Int. J. Mol. Sci. 2017, 18, 2549. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gan, W.; Zou, Y.; Yang, B.; Su, Z.; Deng, J.; Wang, L.; Cai, J. Elevated Levels of Urinary Markers of Oxidative DNA and RNA Damage in Type 2 Diabetes with Complications. Oxid. Med. Cell. Longev. 2016, 2016, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nadagouda, M.N.; Varma, R.S. Green synthesis of silver and palladium nanoparticles at room temperature using coffee and tea extract. Green Chem. 2008, 10, 859. [Google Scholar] [CrossRef]
- Takeoka, H.; Fukui, N.; Sakurai, S.; Nakamura, Y.; Fujii, S. Nanomorphology characterization of sterically stabilized polypyrrole-palladium nanocomposite particles. Polym. J. 2014, 46, 704–709. [Google Scholar] [CrossRef]
- Sheny, D.S.; Philip, D.; Mathew, J. Rapid green synthesis of palladium nanoparticles using the dried leaf of Anacardium occidentale. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 91, 35–38. [Google Scholar] [CrossRef]
- Han, J.; Gurunathan, S.; Jeong, J.-K.; Choi, Y.-J.; Kwon, D.-N.; Park, J.-K.; Kim, J.-H. Oxidative stress mediated cytotoxicity of biologically synthesized silver nanoparticles in human lung epithelial adenocarcinoma cell line. Nanoscale Res. Lett. 2014, 9, 459. [Google Scholar] [CrossRef]
- Murdock, R.C.; Braydich-Stolle, L.; Schrand, A.M.; Schlager, J.J.; Hussain, S.M. Characterization of Nanomaterial Dispersion in Solution Prior to In Vitro Exposure Using Dynamic Light Scattering Technique. Toxicol. Sci. 2008, 101, 239–253. [Google Scholar] [CrossRef]
- Qiu, X.F.; Xu, J.Z.; Zhu, J.M.; Zhu, J.J.; Xu, S.; Chen, H.Y. Controllable synthesis of palladium nanoparticles via a simple sonoelectrochemical method. J. Mater. Res. 2003, 18, 1399–1404. [Google Scholar] [CrossRef]
- Iavicoli, I.; Farina, M.; Fontana, L.; Lucchetti, D.; Leso, V.; Fanali, C.; Cufino, V.; Boninsegna, A.; Leopold, K.; Schindl, R.; et al. In vitro evaluation of the potential toxic effects of palladium nanoparticles on fibroblasts and lung epithelial cells. Toxicol. Vitr. 2017, 42, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Lechon, M.J.; O’Connor, E.; Castell, J.V.; Jover, R. Sensitive Markers Used to Identify Compounds That Trigger Apoptosis in Cultured Hepatocytes. Toxicol. Sci. 2002, 65, 299–308. [Google Scholar] [CrossRef]
- Tee, J.K.; Ong, C.N.; Bay, B.H.; Ho, H.K.; Leong, D.T. Oxidative stress by inorganic nanoparticles. Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology 2016, 8, 414–438. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.P.; Xia, Q.; Hwang, H.-M.; Ray, P.C.; Yu, H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014, 22, 64–75. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Xia, Y.; Niu, P.; Jiang, L.; Duan, J.; Yu, Y.; Zhou, X.; Li, Y.; Sun, Z. Silica nanoparticles induce oxidative stress, inflammation, and endothelial dysfunction in vitro via activation of the MAPK/Nrf2 pathway and nuclear factor-κB signaling. Int. J. Nanomedicine 2015, 10, 1463–1477. [Google Scholar] [CrossRef]
- Novotna, B.; Herynek, V.; Rossner, P.; Turnovcova, K.; Jendelova, P.; Jendelova, P. The effects of grafted mesenchymal stem cells labeled with iron oxide or cobalt-zinc-iron nanoparticles on the biological macromolecules of rat brain tissue extracts. Int. J. Nanomedicine 2017, 12, 4519–4526. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Yan, Q.; Shen, W.; Gurunathan, S. Trichostatin A Enhances the Apoptotic Potential of Palladium Nanoparticles in Human Cervical Cancer Cells. Int. J. Mol. Sci. 2016, 17, 1354. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Combination of palladium nanoparticles and tubastatin-A potentiates apoptosis in human breast cancer cells: A novel therapeutic approach for cancer. Int. J. Nanomedicine 2017, 12, 6503–6520. [Google Scholar] [CrossRef]
- Marí, M.; Morales, A.; Colell, A.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial glutathione, a key survival antioxidant. Antioxid. Redox Signal. 2009, 11, 2685–2700. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, N.; Zhu, M.; Lu, J.; Zhong, H.; Xue, X.; Guo, S.; Li, M.; Wei, X.; Tao, Y.; et al. TiO2 nanoparticles cause mitochondrial dysfunction, activate inflammatory responses, and attenuate phagocytosis in macrophages: A proteomic and metabolomic insight. Redox Biol. 2018, 15, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Chen, Q.; Ding, T.; Sun, J. SiO2 nanoparticle-induced impairment of mitochondrial energy metabolism in hepatocytes directly and through a Kupffer cell-mediated pathway in vitro. Int. J. Nanomedicine 2014, 9, 2891–2903. [Google Scholar] [PubMed]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Jänicke, R.U. MCF-7 breast carcinoma cells do not express caspase-3. Breast Cancer Res. Treat. 2009, 117, 219–221. [Google Scholar] [CrossRef]
- Takaki, K.; Higuchi, Y.; Hashii, M.; Ogino, C.; Shimizu, N. Induction of apoptosis associated with chromosomal DNA fragmentation and caspase-3 activation in leukemia L1210 cells by TiO2 nanoparticles. J. Biosci. Bioeng. 2014, 117, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Sierra, M.I.; Rubio, L.; Bayón, G.F.; Cobo, I.; Menendez, P.; Morales, P.; Mangas, C.; Urdinguio, R.G.; Lopez, V.; Valdes, A.; et al. DNA methylation changes in human lung epithelia cells exposed to multi-walled carbon nanotubes. Nanotoxicology 2017, 11, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zhang, J.; Hu, Q.; Xu, M.; Chen, Y.; Hu, G.; Zhao, M.; Liu, S. Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials 2015, 70, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Igaz, N.; Kovács, D.; Rázga, Z.; Kónya, Z.; Boros, I.M.; Kiricsi, M. Modulating chromatin structure and DNA accessibility by deacetylase inhibition enhances the anti-cancer activity of silver nanoparticles. Colloids Surf. B Biointerfaces 2016, 146, 670–677. [Google Scholar] [CrossRef]
- Landa, P.; Vankova, R.; Andrlova, J.; Hodek, J.; Marsik, P.; Storchova, H.; White, J.C.; Vanek, T. Nanoparticle-specific changes in Arabidopsis thaliana gene expression after exposure to ZnO, TiO2, and fullerene soot. J. Hazard. Mater. 2012, 241–242, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.Q.; Costa-Pinheiro, P.; Almeida-Rios, D.; Graça, I.; Monteiro-Reis, S.; Simões-Sousa, S.; Carneiro, I.; Sousa, E.J.; Godinho, M.I.; Baltazar, F.; et al. SMYD3 contributes to a more aggressive phenotype of prostate cancer and targets Cyclin D2 through H4K20me3. Oncotarget 2015, 6, 13644–13657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Su, L.; Lei, Y.; Liu, X.; Zhang, Y.; Liu, X. DDIT3 and KAT2A Proteins Regulate TNFRSF10A and TNFRSF10B Expression in Endoplasmic Reticulum Stress-mediated Apoptosis in Human Lung Cancer Cells. J. Biol. Chem. 2015, 290, 11108–11118. [Google Scholar] [CrossRef] [Green Version]




| Name of the Sample PdNPs (Concentration (µg/mL)) | Hydrodynamic Size in Dulbecco’s Modified Eagle’s Medium (DMEM) | Zeta Potential in Dulbecco’s Modified Eagle’s Medium (DMEM) |
|---|---|---|
| 1 | 14 | −15.4 ± 1.3 |
| 2 | 13 | −17.3 ± 2.2 |
| 3 | 12 | −18.8 ± 1.9 |
| 4 | 11 | −19.5 ± 2.7 |
| 5 | 11 | −21.8 ± 3.3 |
| Phase | Control | PdNPs | Doxorubicin | Pd(IV) ion |
|---|---|---|---|---|
| GO/G1 | 42.1 ± 2.0 | 58.1 ± 1.8 * | 60.2 ± 2.2 * | 71.5 ± 2.4 * |
| S | 45.7 ± 2.6 | 34.0 ± 1.5 * | 35.1 ± 1.1 * | 24 ± 1.1 * |
| G2/M | 8.5 ± 0.5 | 4.5 ± 0.1 * | 1.3 ± 0.1 * | 0.8 ± 0.2 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurunathan, S.; Qasim, M.; Park, C.H.; Arsalan Iqbal, M.; Yoo, H.; Hwang, J.H.; Uhm, S.J.; Song, H.; Park, C.; Choi, Y.; et al. Cytotoxicity and Transcriptomic Analyses of Biogenic Palladium Nanoparticles in Human Ovarian Cancer Cells (SKOV3). Nanomaterials 2019, 9, 787. https://doi.org/10.3390/nano9050787
Gurunathan S, Qasim M, Park CH, Arsalan Iqbal M, Yoo H, Hwang JH, Uhm SJ, Song H, Park C, Choi Y, et al. Cytotoxicity and Transcriptomic Analyses of Biogenic Palladium Nanoparticles in Human Ovarian Cancer Cells (SKOV3). Nanomaterials. 2019; 9(5):787. https://doi.org/10.3390/nano9050787
Chicago/Turabian StyleGurunathan, Sangiliyandi, Muhammad Qasim, Chan Hyeok Park, Muhammad Arsalan Iqbal, Hyunjin Yoo, Jeong Ho Hwang, Sang Jun Uhm, Hyuk Song, Chankyu Park, Youngsok Choi, and et al. 2019. "Cytotoxicity and Transcriptomic Analyses of Biogenic Palladium Nanoparticles in Human Ovarian Cancer Cells (SKOV3)" Nanomaterials 9, no. 5: 787. https://doi.org/10.3390/nano9050787







