Dietary Nutrients, Proteomes, and Adhesion of Probiotic Lactobacilli to Mucin and Host Epithelial Cells
Abstract
:1. Introduction
2. Gastrointestinal Tract, Mucins, and Lactobacilli
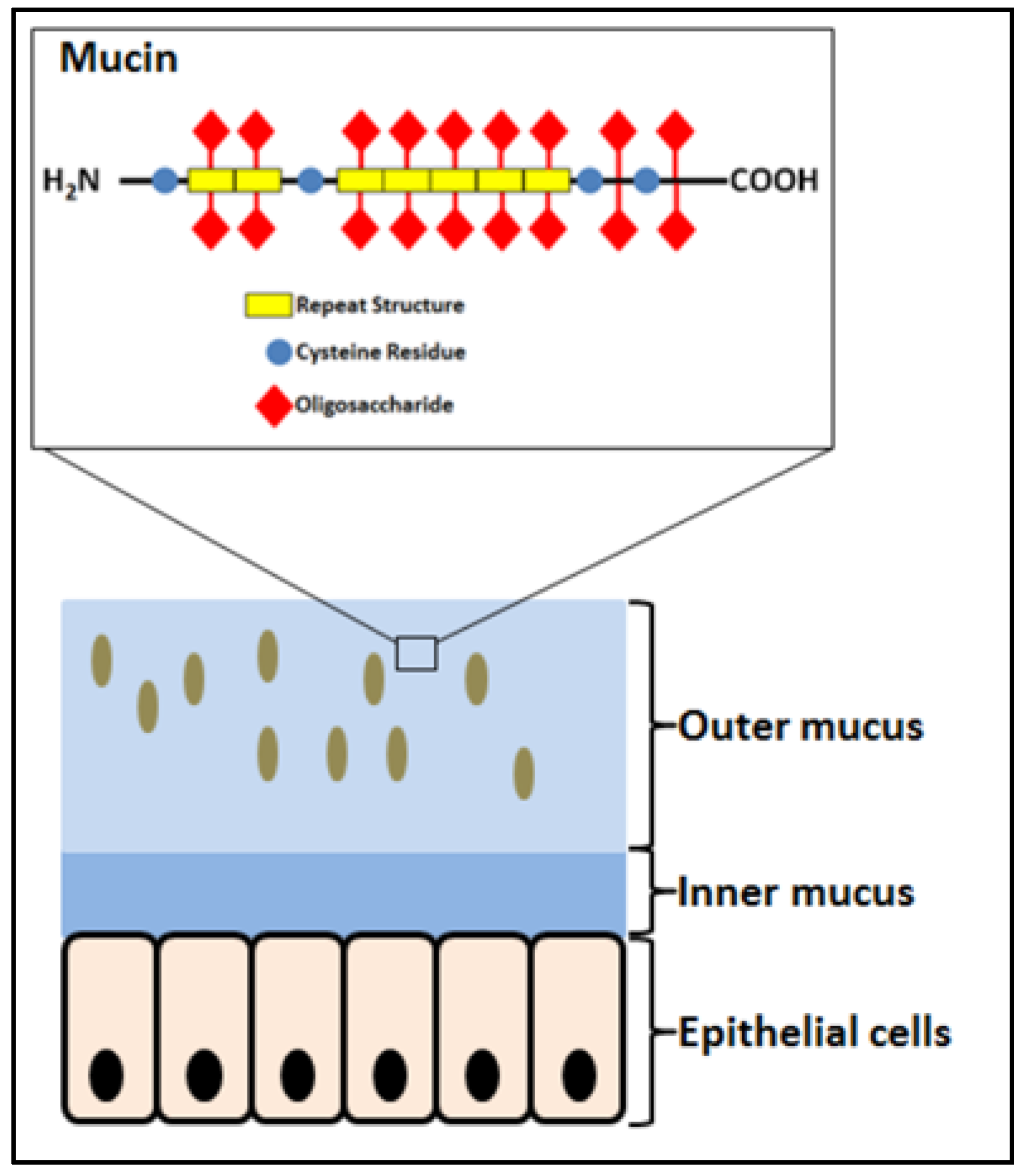
2.1. Mucosa and Mucins
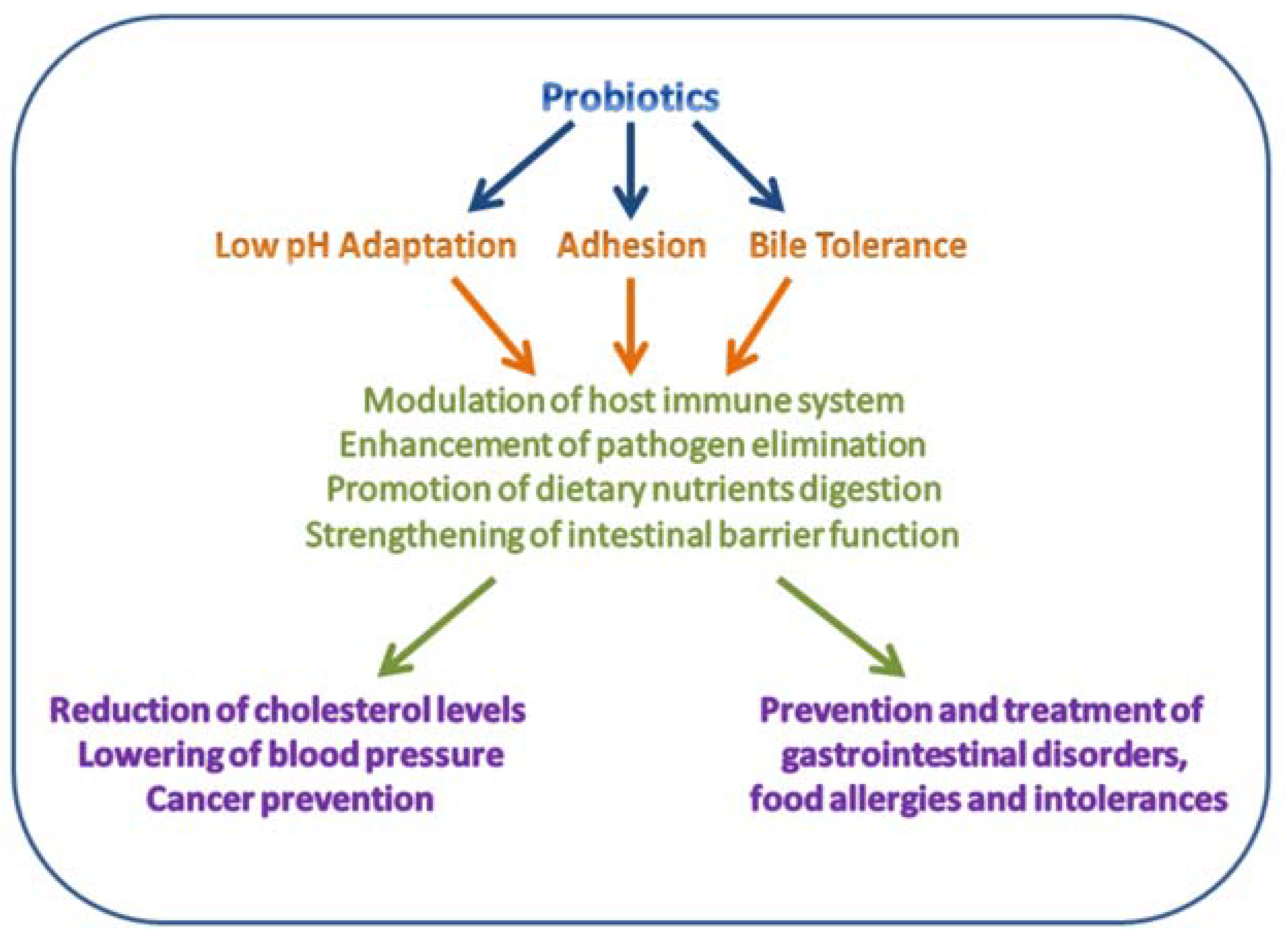
2.2. Probiotic Lactobacilli
3. Lactobacillus Subproteome Analyses
4. Adhesion Factors of Lactobacilli
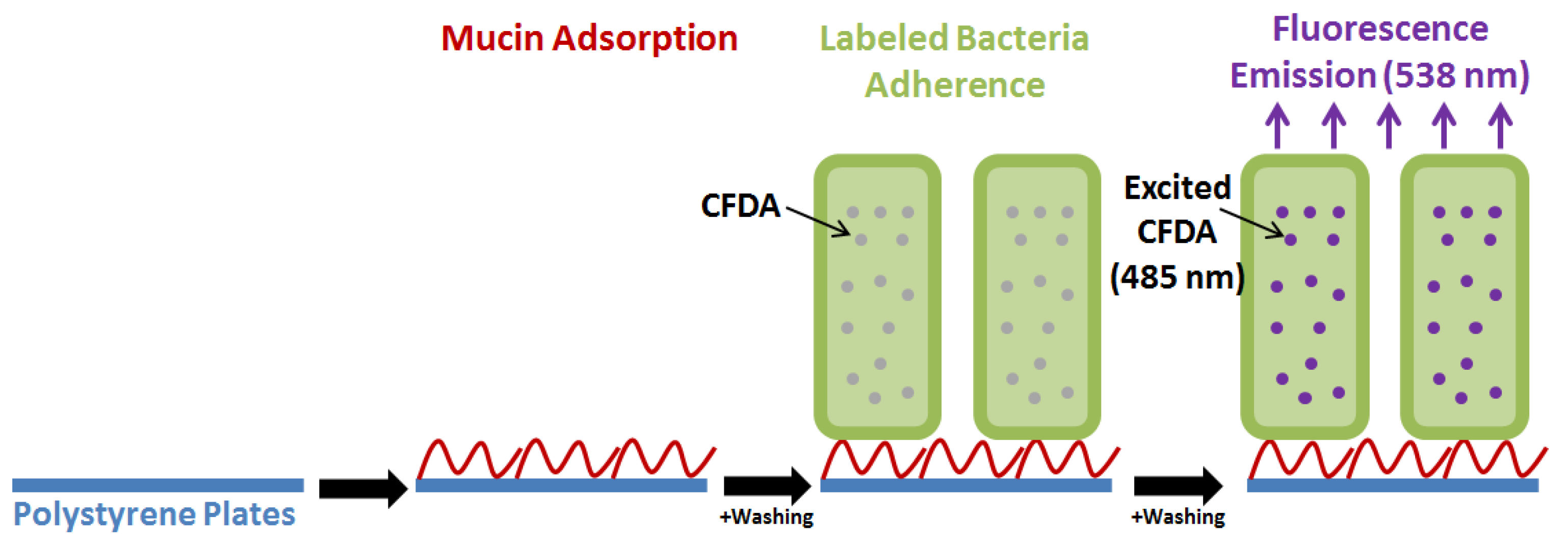
4.1. Assaying Bacterial Adhesion In Vitro
4.2. Identification of Surface Proteins in Lactobacillus acidophilus NCFM
4.3. Some Adhesive Surface Proteins Identified in Other Probiotic Lactobacilli
4.4. Identification of Moonlighting Proteins in Probiotic Lactobacilli
5. Effect of Compounds in the Diet on Adhesion of Lactobacilli
5.1. Prebiotics
5.2. Synbiotics
6. Dietary Nutrients-Lactobacillus Interactions Characterized by Adherence and Proteome Analyses
7. Conclusions and Future Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Roberfroid, M.B. A European consensus of scientific concepts of functional foods. Nutrition 2000, 16, 689–691. [Google Scholar] [CrossRef]
- Voreades, N.; Kozil, A.; Weir, T.L. Diet and the development of the human intestinal microbiome. Front. Microbiol. 2014, 5, 494. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; Kleerebezeem, M.; Brummer, R.-J.; Cani, P.D.; Mercenier, A.; MacDonald, T.T.; Garcia-Ródenas, C.L.; Wells, J.M. Can probiotics modulate human disease by impacting intestinal barrier function? Br. J. Nutr. 2017, 117, 93–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markowiak, P.; Slizewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Capozzi, V.; Spano, G.; Fiocco, D. The potential of lactic acid bacteria to colonize bionic and abiotic surfaces and the investigation of their interactions. Appl. Microbiol. Biotechnol. 2017, 101, 2641–2657. [Google Scholar] [CrossRef] [PubMed]
- Deepika, G.; Charalampopoulos, D. Surface and adhesion properties of Lactobacilli. Adv. Appl. Microbiol. 2010, 40, 127–152. [Google Scholar] [CrossRef]
- Buck, B.; Altermann, E.; Svingerud, T.; Klaenhammer, T.R. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2005, 71, 8344–8351. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.J.; Klaenhammer, T.R. Functional and phenotypic characterization of a protein from L. acidophilus involved in cell morphology, stress tolerance and adhesions to intestinal cells. Microbiology 2010, 156, 3360–3367. [Google Scholar] [CrossRef] [PubMed]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.R.; O’Flaherty, S.; Goh, Y.J.; Carroll, I.; Barrangou, R.; Klaenhammer, T.R. The S-layer associated serine protease homologue PrtX impacts cell surface-mediated microbe-host interactions of Lactobacillus acidophilus NCFM. Front. Microbiol. 2017, 8, 1185. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Selle, K.; O’Flaherty, S.; Goh, Y.J.; Klaenhammer, T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 2013, 159, 2269–2282. [Google Scholar] [CrossRef] [PubMed]
- Amenyogbe, N.; Kollmann, T.R.; Ben-Othman, R. Early-life host-microbiome interface: The key frontier for immune development. Front. Pediatr. 2017, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Park, W. Gut microbiomes and their metabolites shape human and animal health. J. Microbiol. 2018, 56, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Bansil, R.; Turner, B.S. Mucin structure, aggregation, physiological functions and biomedical applications. Curr. Opin. Colloid Interface Sci. 2006, 11, 164–170. [Google Scholar] [CrossRef]
- Van Tassell, M.L.; Miller, M.J. Lactobacillus adhesion to mucus. Nutrients 2011, 3, 613–636. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K. Is it feasible to control pathogen infection by competitive binding of probiotics to the host? Virulence 2017, 8, 1502–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etzold, S.; Juge, N. Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 2014, 28, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Bäckström, M.; Ambort, D.; Thomsson, E.; Johansson, M.E.V.; Hansson, G.C. Increased understanding of the biochemistry and biosynthesis of MUC2 and other gel-forming mucins through the recombinant expression of their protein domains. Mol. Biotechnol. 2013, 54, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Slover, C.M.; Danziger, L. Lactobacillus: A review. Clin. Microbiol. Newsl. 2008, 30, 23–27. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Dunlap, P.V.; Clark, D.P. Bacteria: Gram-positive and other bacteria. In Brock: Biology of Microorganisms; Madigan, M.T., Martinko, J.M., Dunlap, P.V., Clark, D.P., Eds.; Pearson Benjamin Cummings: San Francisco, CA, USA, 2009; pp. 446–486. ISBN 978-3-319-48325-2. [Google Scholar]
- Sanders, M.E. How do we know when something called “probiotic” is really a probiotic? A guideline for consumers and health care professionals. Funct. Food Rev. 2009, 1, 3–12. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.Q.A.; Altermann, E.; Goh, Y.J.; Tallon, R.; Sanozky-Dawes, R.B.; Pfeiler, E.A.; O’Flaherty, S.; Buck, B.L.; Dobson, A.; Duong, T.; et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 2008, 74, 4610–4625. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.; Maqsood, S.; Masud, T.; Ahmad, A.; Sohail, A.; Momin, A. Lactobacillus acidophilus: Charcaterization of the species and application in food production. Crit. Rev. Food Sci. Nutr. 2014, 54, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Vignolles, M.-L.; Chen, X.D.; Le Loir, Y.; Jan, G.; Schuck, P.; Jeantet, R. Spray drying of probiotics and other food-grade bacteria: A review. Trends Food Sci. Technol. 2017, 63, 1–17. [Google Scholar] [CrossRef]
- Collins, M.D.; Gibson, G.R. Probiotics, prebiotics, and synbiotics: Approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 1999, 69, 1052S–1057S. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An overview of beneficial effects. Antonie van Leeuwenhoek 2002, 82, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, R.A.; Mazzeo, M.F. Molecular mechanisms of probiotic action: A proteomic perspective. Curr. Opin. Microbiol. 2012, 15, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Muyyarikkandy, M.S.; Amalaradjou, M.A. Lactobacillus bulgaricus, Lactobacillus rhamnosus and Lactobacillus paracasei attenuate Salmonella enteritidis, Salmonella Heidelberg and Salmonella typhimurium colonization and virulence gene expression in vitro. Int. J. Mol. Sci. 2017, 18, 2381. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Leonhard, M.; Moser, D.; Schneider-Stickler, B. Inhibition activity of Lactobacilli supernatant against fungal-bacterial multispecies biofilm on silicone. Microb. Pathog. 2017, 113, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, B.; López, P.; González-Rodrígez, I.; Suárez, A.; Margolles, A.; Urdaci, M.C. A flagellin-producing Lactococcus strain: Interaction with mucin and enteropathogens. FEMS Microbiol. 2011, 318, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Son, S.-H.; Jeon, H.-L.; Yang, S.-J.; Lee, N.-K.; Paik, H.-D. In vitro characterization of Lactobacillus brevis KU15006, an isolate from kimchi, reveals anti-adhesion activity against foodborne pathogens and antidiabetic properties. Microb. Pathog. 2017, 112, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Buntin, N.; de Vos, W.M.; Hongpattarakare, T. Variation of mucin adhesion, cell surface characteristics, and molecular mechanisms among Lactobacillus plantarum isolated from different habitats. Appl. Microbiol. Biotechnol. 2017, 101, 7663–7674. [Google Scholar] [CrossRef] [PubMed]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Lactobacillus fermentum 3872 as a potential tool for combatting Campylobacter jejuni infections. Virulence 2017, 8, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Cremon, C.; Guglielmetti, S.; Gargari, G.; Taverniti, V.; Castellazzi, A.M.; Valsecchi, C.; Tagliacarne, C.; Fiore, W.; Bellini, M.; Bertani, L.; et al. Effect of Lactobacillus paracasei CNCM I-1572 on symptoms, gut microbiota, short chain fatty acids, and immune activation in patients with irritable bowel syndrome: A pilot randomized clinical trial. United European Gastroenterol. J. 2018, 6, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Madden, J.A.J.; Hunter, J.O. A review of the role of the gut microflora in irritable bowel syndrome and the effects of probiotics. Br. J. Nutr. 2002, 88, s67–s72. [Google Scholar] [CrossRef] [PubMed]
- Hempel, S.; Newberry, S.J.; Maher, A.R.; Wang, Z.; Miles, J.N.V.; Shanman, R.; Johnsen, B.; Shekelle, P.G. Probiotics for the prevention and treatment of antibiotic-associated diarrhea. JAMA 2012, 307, 1959–1969. [Google Scholar] [CrossRef] [PubMed]
- Pajarillo, E.A.B.; Kim, S.H.; Valeriano, V.D.; Lee, J.Y.; Kang, D.-K. Proteomic view of the crosstalk between Lactobacillus mucosae and intestinal epithelial cells in co-culture revealed by Q Exactive-based quantitative proteomics. Front. Microbiol. 2017, 8, 2459. [Google Scholar] [CrossRef] [PubMed]
- Klotz, C.; O’Flaherty, S.; Goh, Y.J.; Barrangou, R. Investigating the effects of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front. Microbiol. 2017, 8, 2174. [Google Scholar] [CrossRef] [PubMed]
- Van de Guchte, M.; Chaze, T.; Jan, G.; Mistou, M.-Y. Properties of probiotic bacteria explored by proteomic approaches. Curr. Opin. Microbiol. 2012, 15, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Celebioglu, H.U.; Svensson, B. Exo- and surface proteomes of the probiotic bacterium Lactobacillus acidophilus NCFM. Proteomics 2017, 17, 1700019. [Google Scholar] [CrossRef] [PubMed]
- Ashida, N.; Yanagihara, S.; Shinoda, T.; Yamamoto, N. Characterization of adhesive molecule with affinity for Caco-2 cells in Lactobacillus acidophilus by proteome analysis. J. Biosci. Bioeng. 2011, 112, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Celebioglu, H.U.; Ejby, M.; Majumder, A.; Købler, C.; Goh, Y.J.; Thorsen, K.; Schmidt, B.; O’Flaherty, S.; Abou Hachem, M.; Lahtinen, S.J.; et al. Differential proteome and cellular adhesion analyses of the probiotic bacterium Lactobacillus acidophilus NCFM grown on raffinose—An emerging prebiotic. Proteomics 2016, 16, 1361–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celebioglu, H.U.; Olesen, S.V.; Prehn, K.; Lahtinen, S.J.; Brix, S.; Abou Hachem, M.; Svensson, B. Mucin- and carbohydrate-stimulated adhesion and subproteome changes of the probiotic bacterium Lactobacillus acidophilus NCFM. J. Proteomics 2017, 163, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Espino, E.; Koskenniemi, K.; Mato-Rodriguez, L.; Nyman, T.A.; Reunanen, J.; Koponen, J.; Ohman, T.; Siljamaki, P.; Alatossava, T.; Varmanen, P.; et al. Uncovering surface-exposed antigens of Lactobacillus rhamnosus by cell shaving proteomics and two-dimensional immunoblotting. J. Proteome Res. 2015, 14, 1010–1024. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.R.; Hymes, J.; Sanozky-Dawnes, R.; Henriksen, E.D.; Barrangou, R.; Klaenhammer, T.R. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl. Environ. Microbiol. 2016, 82, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Do Carmo, F.L.R.; Rabah, H.; Huang, S.; Gaucher, F.; Deplanche, M.; Dutertre, S.; Jardin, J.; Le Loir, Y.; Azevedo, V.; Jan, G. Propionibacterium freudenreichii surface protein SlpB is involved in adhesion to intestinal HT-29 cells. Front. Microbiol. 2017, 8, 1033. [Google Scholar] [CrossRef] [PubMed]
- Bentley-Hewitt, K.L.; Narbad, A.; Majsak-Newman, G.; Philo, M.R.; Lund, E.K. Lactobacilli survival and adhesion to colonic epithelial cell lines is dependent on long chain fatty acid exposure. Eur. J. Lipid Sci. Technol. 2017, 119, 1700062. [Google Scholar] [CrossRef]
- Wasko, A.T.; Polak-Berecka, M.; Paduch, R.; Józwiak, K. The effect of moonlighting proteins on the adhesion and aggregation ability of Lactobacillus helveticus. Anaerobe 2014, 30, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, E.; Horvatovich, P.; Machioni, E.; Aoude-Werner, D.; Sanz, Y.; Ennahar, S. 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis 2009, 30, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Leteurtre, T.; Gouyer, V.; Rousseau, K.; Moreau, O.; Barbat, A.; Swallow, D.; Lesuffleur, T. Differential mucin expression in colon carcinoma HT-29 clones with variable resistance to 5-fluorouracil and methotrexate. Biol. Cell 2004, 96, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, M.; Robineleon, S.; Appay, M.D.; Kedinger, M.; Triadou, N.; Dussaulx, E.; Lacroix, B.; Simonassmann, P.; Haffen, K.; Fogh, J.; et al. Enterocyte-like differentiation and polarization of the human-colon carcinoma cell-line Caco-2 in culture. Biol. Cell 1983, 47, 323–340. [Google Scholar]
- Spacova, I.; Lievens, E.; Verhoeven, T.; Steenackers, H.; Vanderleyden, J.; Lebeer, S.; Petrova, M.I. Expression of fluorescent proteins in Lactobacillus rhamnosus to study host−microbe and microbe−microbe interactions. Microb. Biotechnol. 2018, 11, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.K.; Shah, K.R.; Pappachan, A.; Gupta, S.; Singh, D.D. Cloning, expression and characterisation of mucin binding GAPDH from Lactobacillus acidophilus. Int. J. Biol. Macromol. 2016, 91, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Gallego, J.; Arroyo-López, F.N.; Rantsiou, K.; Jiménez-Díaz, R.; Garrido-Fernández, A.; Cocolin, L. Screening of lactic acid bacteria isolated from fermented table olives with probiotic potential. Food Res. Intl. 2013, 50, 135–142. [Google Scholar] [CrossRef]
- Campaniello, D.; Speranza, B.; Petruzzi, L.; Bevilacqua, A.; Corbo, M.R. How to routinely assess transition, adhesion and survival of probiotics into the gut: A case study of propionibacteria. Int. J. Food Sci. Technol. 2018, 53, 484–490. [Google Scholar] [CrossRef]
- Bujnakova, D.; Kmet, V. Aggregation of animal Lactobacilli with O157 enterohemorrhagic Escherichia coli. J. Vet. Med. Ser. B 2002, 49, 152–154. [Google Scholar] [CrossRef]
- Goh, Y.J.; Klaenhammer, T.R. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2010, 76, 5005–5012. [Google Scholar] [CrossRef] [PubMed]
- Hymes, J.P.; Johnson, B.R.; Barrangou, R.; Klaenhammer, T.R. Functional analysis of an S-layer associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2016, 82, 2676–2685. [Google Scholar] [CrossRef] [PubMed]
- Bengoa, A.A.; Zavala, L.; Carasi, P.; Trejo, S.A.; Bronsoms, S.; de los Angeles Serradell, M.; Garrote, G.L.; Abraham, A.G. Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res. Intl. 2018, 103, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Boekhorst, J.; Helmer, Q.; Kleerebezem, M.; Siezen, R.J. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 2006, 152, 273–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etzold, S.; Kober, O.L.; MacKenzie, D.A.; Tailford, L.E.; Gunning, A.P.; Walshaw, J.; Hemmings, A.M.; Juge, N. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 2014, 16, 888–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hytönen, J.; Hataja, S.; Finne, J. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect. Immun. 2003, 71, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Hytönen, J.; Hataja, S.; Finne, J. Use of flow cytometry for the adhesion analysis of Streptococcus pyogenes mutant strain to epithelial cells: Investigation of the possible role of surface pullulanase and cysteine protease, and the transcriptional regulator Rgg. BMC Microbiol. 2006, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Aryantini, N.P.D.; Kondoh, D.; Nishiyama, K.; Yamamoto, Y.; Mukai, T.; Sujaya, I.N.; Urashima, T.; Fukuda, K. Anchorless cell surface proteins function as laminin-binding adhesins in Lactobacillus rhamnosus FSMM22. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Okochi, M.; Sugita, T.; Asai, Y.; Tanaka, M.; Honda, H. Screening of peptides associated with adhesion and aggregation of Lactobacillus rhamnosus GG in vitro. Biochem. Eng. J. 2017, 128, 178–185. [Google Scholar] [CrossRef]
- Kainulainen, V.; Korhonen, T.K. Dancing to another tune—Adhesive moonlighting proteins in bacteria. Biology 2014, 3, 178–204. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Protein moonlighting: What is it, and why is it important. Phil. Trans. R. Soc. B. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, C.; Flores, C.-L.; Gancedo, J.M. The expanding landscape of moonlighting proteins in yeasts. Micromol. Mol. Biol. Rev. 2016, 80, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Mani, M.; Chen, C.; Amblee, V.; Liu, H.; Mathur, T.; Zwicke, G.; Zabad, S.; Patel, B.; Thakkar, J.; Jeffery, C.J. MoonProt: A database for proteins that are known to moonlight. Nucleic Acids Res. 2015, 43, D277–D282. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.-F.; Zhang, L.-J.; Pang, X.-H.; Gu, X.-X.; Abdelazez, A.; Liang, Y.; Sun, S.-R.; Meng, X.-C. Complete genome sequence of bacteriocin-producing Lactobacillus plantarum KLDS1.0391, a probiotic strain with gastrointestinal tract resistance and adhesion to the intestinal epithelial cell. Genomics 2017, 109, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Ramiah, K.; van Reenen, C.A.; Dicks, L.M. Surface-bound proteins of Lactobacillus plantarum 423 that contribute to adhesion of Caco-2 cells and their role in competitive exclusion and displacement of Clostridium sporogenes and Enterococcus faecilis. Res. Microbiol. 2008, 159, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [PubMed]
- Gibson, G.R.; Probert, H.M.; Van Loo, J.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.J.; Klaenhammer, T.R. Genetic mechanisms of prebiotic oligosaccharide metabolism in probiotic microbes. Ann. Rev. Food Sci. Technol. 2015, 6, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Li, D.; Zhao, J.; Liu, X.; Gu, Z.; Chen, Y.Q.; Zhang, H.; Chen, W. In vitro fermentation of fructooligosaccharides with human gut bacteria. Food Funct. 2015, 6, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Scott, K.P.; Rastall, R.A.; Tuohy, M.M.; Hotchkiss, A.; Dubert-Ferrandon, A.; Garau, M.; Murphy, E.F.; Saulnier, D.; Loh, G.; et al. Dietary prebiotics: Current status and new definition. Food Sci Technol Bull Funct Foods 2010, 7, 1–19. [Google Scholar] [CrossRef]
- Kolida, S.; Gibson, G.R. Synbiotics in health and disease. Annu. Rev. Food Sci. Technol. 2011, 2, 373–393. [Google Scholar] [CrossRef] [PubMed]
- Krumbeck, J.A.; Water, J.; Hutkins, R.W. Synbiotics for improved human health: Recent developments, challenges, and opportunities. Annu. Rev. Food. Sci. Technol. 2018, 9, 451–479. [Google Scholar] [CrossRef] [PubMed]
- Kimoto-Nira, H.; Yamasaki, S.; Sasaki, K.; Moriya, N.; Takenaka, A.; Suzuki, C. New lactic acid bacterial strains from traditional Mongolian fermented milk products have altered adhesion to porcine gastric mucin depending on the carbon source. Anim. Sci. J. 2015, 86, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Cardona, F.; Andrés-Lacueva, C.; Tulipani, S.; Tinahones, F.J.; Queipo-Ortuno, M.I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutri. Biochem. 2013, 24, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- De Llano, D.G.; Gill-Sanchez, I.; Esteban-Fernande, A.; Ramos, A.M.; Fernandez-Diaz, M.; Cueva, C.; Moreno-Arribas, M.V.; Bartolome, B. Reciprocal beneficial effects between wine polyphenols and probiotics: An exploratory study. Eur. Food Res. Technol. 2017, 243, 531–538. [Google Scholar] [CrossRef]
- Celebioglu, H.U.; Delsoglio, M.; Brix, S.; Pessione, E.; Svensson, B. Plant polyphenols stimulate adhesion to intestinal mucosa and induce proteome changes in the probiotic Lactobacillus acidophilus NCFM. Mol. Nutr. Food Res. 2018, 62, 1700638. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Jespersen, L. Transcriptional analysis of genes associated with stress and adhesion in Lactobacillus acidophilus NCFM during passage through an in vitro gastrointestinal tract model. J. Mol. Microbiol. Biotechnol. 2010, 18, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A.; Sultan, A.; Jersie-Christensen, R.R.; Ejby, M.; Schmidt, B.; Lahtinen, S.J.; Jacobsen, S.; Svensson, B. Proteome reference map of Lactobacillus acidophilus NCFM and quantitative proteomics towards understanding the prebiotic action of lactitol. Proteomics 2011, 11, 3470–3481. [Google Scholar] [CrossRef] [PubMed]
- Van Zanten, G.C.; Sparding, N.; Majumder, A.; Lahtinen, S.J.; Svensson, B.; Jacobsen, S. The acidic and alkaline differential proteomes of Lactobacillus acidophilus NCFM grown on cellobiose shows up-regulation of two β-glycoside hydrolases and proteins involved in adherence. BioMed Res. Int. 2015, 2015, 347216. [Google Scholar] [CrossRef] [PubMed]
- Van Zanten, G.; Krych, L.; Röytiö, H.; Forssten, S.; Lahtinen, S.; Al-Soud, W.; Sørensen, S.; Svensson, B.; Jespersen, L.; Jakobsen, M. Synbiotic Lactobacillus acidophilus NCFM and cellobiose does not affect human gut bacterial diversity but increases abundance of lactobacilli, bifidobacteria and branched-chain fatty acids: A randomized, double-blinded cross-over trial. FEMS Microbiol. Ecol. 2014, 90, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Van Zanten, G.C.; Knudsen, A.; Röytiö, H.; Forssten, S.; Lawther, M.; Blennow, A.; Lahtinen, S.J.; Jakobsen, M.; Svensson, B.; Jespersen, L. The effect of selected synbiotics on microbial composition and short-chain fatty acid production in a model system of the human colon. PLoS ONE 2012, 7, e47212. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; López, P.; Capozzi, V.; Fernández de Palencia, P.; Duenas, M.T.; Spano, G.; Fiocco, D. Beta-glucans improve growth, viability and colonization of probiotic microorganisms. Int. J. Mol. Sci. 2012, 13, 6026–6039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allonsius, C.N.; van den Broek, M.F.L.; De Boeck, I.; Kiekens, S.; Oerlemans, E.F.M.; Kiekens, F.; Foubert, K.; Vandenheuvei, D.; Cos, P.; et al. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb. Biotechnol. 2017, 10, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
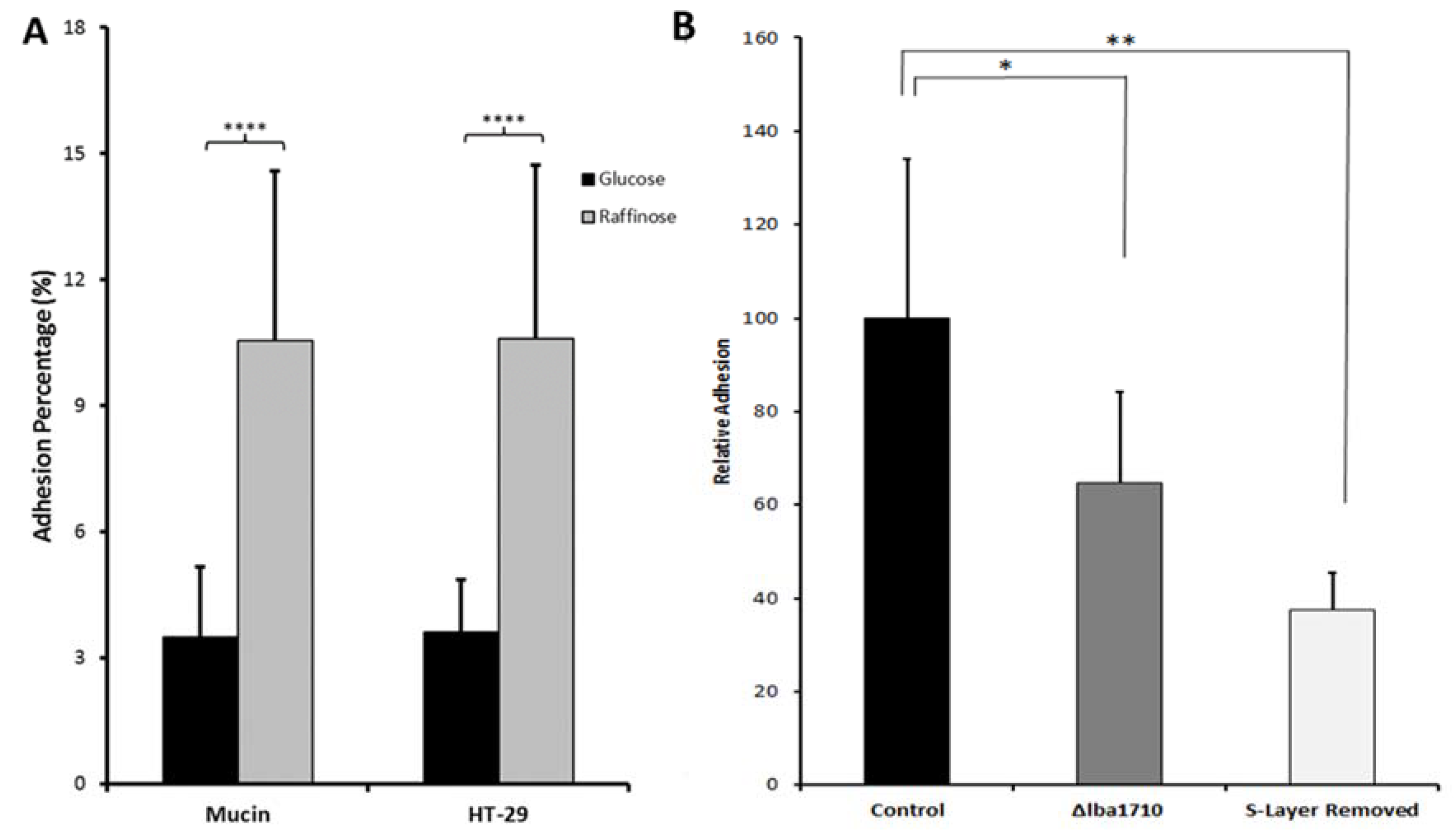

| Carbon Source (1%)/Supplementation | Mucin d (Fold-Increase) | HT-29 d (Fold-Increase) |
|---|---|---|
| Raffinose a | 3 (p < 0.0001) | 3 (p < 0.0001) |
| Cellobiose b | 2.5 (p < 0.05) | 2.5 (p < 0.05) |
| Polydextrose b | 3 (p < 0.05) | 2 (p < 0.05) |
| FOS b | n.d. | 2 (p < 0.05) |
| Glucose/mucin (0.1%) b | n.d. | 2 (p < 0.05) |
| Glucose/resveratrol c | 2.3 (p < 0.01) (100 µg/mL) | 1.4 (p < 0.01) (100 µg/mL) 3.0 (p < 0.001) (250 µg/mL) |
| Glucose/tannic acid c | 0.4 (p < 0.001) (250 µg/mL) | 5.0 (p < 0.001) (100 µg/mL) |
| Glucose/caffeic acid c | 1.3 (p < 0.05) (250 µg/mL) | n.d. |
| Glucose/ferulic acid c | 0.8 (p < 0.01) (100 µg/mL) 1.9 (p < 0.001) (250 µg/mL) 1.2 (p < 0.001) (500 µg/mL) | 1.4 (p < 0.001) (250 µg/mL) |
| Protein Name | Accession Number | Carbon Source and Protein Fold-Change | ||||||
|---|---|---|---|---|---|---|---|---|
| Ra a | Ce b | Mu b | Re c | TA c | CA c | FA c | ||
| Phosphate starvation inducible protein stress-related | YP_193579 | +4.4 | +4.4 | |||||
| Thermostable pullulanase | YP_194553 | +2.3 | +3.5 | |||||
| Elongation factor G M | YP_193213 | +2.1 | +2.4 | −3.5 | −2.0 | |||
| 50S Ribosomal protein L7/L12 | YP_193293 | +2.0 | ||||||
| Pyruvate kinase M | YP_193840 | −1.7 | +1.9 | +2.1 | −1.6 | |||
| Fructose-bis-phosphate aldolase M | YP_194445 | +1.8 | ||||||
| Elongation factor P | YP_194511 | +1.7 | −1.7 | |||||
| 50S Ribosomal protein L22 | YP_193220 | +1.6 | ||||||
| Glutamyl tRNA synthase | YP_193270 | +1.5 | ||||||
| Hypothetical protein LBA1769 | YP_194608 | +1.4 | ||||||
| Aminopeptidase | YP_104682 | +1.4 | ||||||
| S-layer Protein | YP_193101 | +1.3 | ||||||
| Glycoprotein endopeptidase | YP_193310 | −1.4 | +1.3 | |||||
| 6-phophofructokinase M | YP_193839 | −1.4 | ||||||
| Phosphoglycerate kinase M | YP_193605 | −1.4 | −3.1 | |||||
| Molecular chaperone GroEL M | YP_193328 | −1.5 | ||||||
| D-lactate dehydrogenase | YP_192990 | −1.5 | ||||||
| 50S Ribosomal protein L10 | YP_193292 | −1.6 | ||||||
| Trigger factor M | YP_193738 | −1.7 | ||||||
| 50S Ribosomal protein L1 | YP_193283 | −1.8 | ||||||
| ATP synthase FOF1 subunit alpha | YP_193673 | −1.8 | ||||||
| Oligoribonuclease | YP_193337 | −1.8 | ||||||
| Elongation factor Tu M | YP_193737 | −1.8 | −1.8 | |||||
| Lysine tRNA ligase | YP_193205 | −1.9 | ||||||
| L-lactate dehydrogenase | YP_193195 | −1.6 | ||||||
| Elongation factor Ts | YP_194131 | −2.0 | ||||||
| Triose phosphate isomerase M | YP_193606 | −2.0 | −2.0 | −1.7 | ||||
| Manganese-dependent inorganic pyrophosphatase | YP_194000 | −1.5 | −2.1 | −1.5 | −1.5 | |||
| Aspartate tRNA ligase | YP_193821 | −2.2 | ||||||
| Adenylosuccinate synthase | YP_194721 | −2.3 | −1.5 | −1.5 | ||||
| 30S Ribosomal protein S1 M | YP_193850 | −2.3 | −1.8 | −1.7 | ||||
| Ribonucleoside triphosphate reductase | YP_192977 | −2.3 | ||||||
| Glyceraldehyde-3-p dehydrogenase M (GAPDH) | YP_193579 | −2.4 −1.9 −1.6 | −2.0 −1.5 | −2.0 −1.6 | ||||
| BipAEFTU family GTP-binding protein M | YP_193724 | −2.7 | ||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celebioglu, H.U.; Svensson, B. Dietary Nutrients, Proteomes, and Adhesion of Probiotic Lactobacilli to Mucin and Host Epithelial Cells. Microorganisms 2018, 6, 90. https://doi.org/10.3390/microorganisms6030090
Celebioglu HU, Svensson B. Dietary Nutrients, Proteomes, and Adhesion of Probiotic Lactobacilli to Mucin and Host Epithelial Cells. Microorganisms. 2018; 6(3):90. https://doi.org/10.3390/microorganisms6030090
Chicago/Turabian StyleCelebioglu, Hasan Ufuk, and Birte Svensson. 2018. "Dietary Nutrients, Proteomes, and Adhesion of Probiotic Lactobacilli to Mucin and Host Epithelial Cells" Microorganisms 6, no. 3: 90. https://doi.org/10.3390/microorganisms6030090






