Monitoring and Determining Mitochondrial Network Parameters in Live Lung Cancer Cells
Abstract
:1. Introduction
2. Experimental Section
2.1. Tissue Microarray
2.2. Cell Culture Reagents
2.3. Preparation of BEAS-2B and SCLC Cells for Scanning Electron Microscopy
2.4. Serial Block-Face Scanning Electron Microscopy (SEM)
2.5. Mitochondrial Staining in Live Cells and Confocal Microscopic Imaging
2.6. Integrating Imaging
2.7. Mono-Fractal Analysis
2.8. Multifractal Analysis
2.9. Statistical Analysis
2.10. 3D Reconstruction of SCLC Mitochondria
3. Results
3.1. The Mitochondrial Fission-Associated Protein DRP1 Expression is Increased in SCLC
3.2. Live Cell Imaging to Monitor Mitochondrial Morphology and Dynamics Using MitoMorF
3.3. Mono-Fractal and Multifractal Analysis
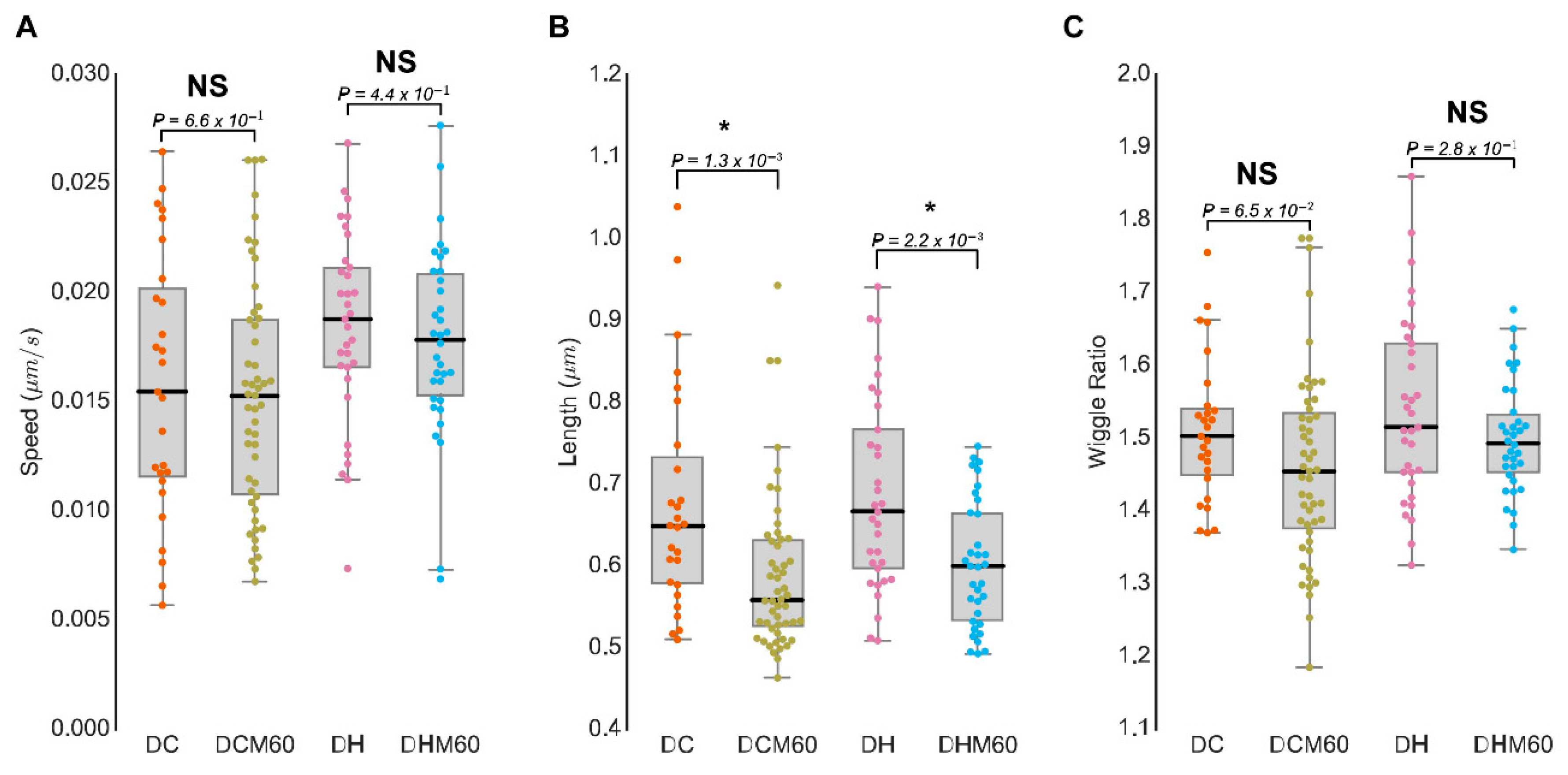
3.4. MitoMorF Can Discern Subtle Differences in Mitochondrial Dynamics
3.5. Three-Dimensional Analysis of Mitochondrial Network to Identify Differences in Mitochondrial Morphology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simula, L.; Nazio, F.; Campello, S. The mitochondrial dynamics in cancer and immune-surveillance. Semin. Cancer Biol. 2017, 47, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Guha, M.; Kashina, A.; Avadhani, N.G. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim. Biophys. Acta 2017, 1858, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.P.; Chipuk, J.E. Mitochondrial dynamics as regulators of cancer biology. Cell. Mol. Life Sci. 2017, 74, 1999–2017. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, A.V.; Hermann, M.; Saks, V.; Hengster, P.; Margreiter, R. The cell-type specificity of mitochondrial dynamics. Int. J. Biochem. Cell Biol. 2009, 41, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Senft, D.; Ronai, Z.A. Regulators of mitochondrial dynamics in cancer. Curr. Opin. Cell Biol. 2016, 39, 43–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wai, T.; Langer, T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef]
- Pendin, D.; Filadi, R.; Pizzo, P. The Concerted Action of Mitochondrial Dynamics and Positioning: New Characters in Cancer Onset and Progression. Front. Oncol. 2017, 7, 102. [Google Scholar] [CrossRef]
- Maycotte, P.; Marin-Hernandez, A.; Goyri-Aguirre, M.; Anaya-Ruiz, M.; Reyes-Leyva, J.; Cortes-Hernandez, P. Mitochondrial dynamics and cancer. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [Green Version]
- Rambold, A.S.; Pearce, E.L. Mitochondrial Dynamics at the Interface of Immune Cell Metabolism and Function. Trends Immunol. 2018, 39, 6–18. [Google Scholar] [CrossRef]
- Trevisan, T.; Pendin, D.; Montagna, A.; Bova, S.; Ghelli, A.M.; Daga, A. Manipulation of Mitochondria Dynamics Reveals Separate Roles for Form and Function in Mitochondria Distribution. Cell Rep. 2018, 23, 1742–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westrate, L.M.; Drocco, J.A.; Martin, K.R.; Hlavacek, W.S.; MacKeigan, J.P. Mitochondrial morphological features are associated with fission and fusion events. PLoS ONE 2014, 9, e95265. [Google Scholar] [CrossRef] [PubMed]
- Giedt, R.J.; Fumene Feruglio, P.; Pathania, D.; Yang, K.S.; Kilcoyne, A.; Vinegoni, C.; Mitchison, T.J.; Weissleder, R. Computational imaging reveals mitochondrial morphology as a biomarker of cancer phenotype and drug response. Sci. Rep. 2016, 6, 32985. [Google Scholar] [CrossRef]
- Harwig, M.C.; Viana, M.P.; Egner, J.M.; Harwig, J.J.; Widlansky, M.E.; Rafelski, S.M.; Hill, R.B. Methods for imaging mammalian mitochondrial morphology: A prospective on MitoGraph. Anal. Biochem. 2018, 552, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Weigert, R.; Sramkova, M.; Parente, L.; Amornphimoltham, P.; Masedunskas, A. Intravital microscopy: A novel tool to study cell biology in living animals. Histochem. Cell Biol. 2010, 133, 481–491. [Google Scholar] [CrossRef]
- Masedunskas, A.; Porat-Shliom, N.; Tora, M.; Milberg, O.; Weigert, R. Intravital microscopy for imaging subcellular structures in live mice expressing fluorescent proteins. J. Vis. Exp. 2013. [Google Scholar] [CrossRef]
- Ouellet, M.; Guillebaud, G.; Gervais, V.; Lupien St-Pierre, D.; Germain, M. A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLoS Comput. Biol. 2017, 13, e1005612. [Google Scholar] [CrossRef]
- Viana, M.P.; Lim, S.; Rafelski, S.M. Quantifying mitochondrial content in living cells. Methods Cell Biol. 2015, 125, 77–93. [Google Scholar]
- Liesa, M.; Palacín, M.; Zorzano, A. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef]
- Ma, P.C.; Jagadeeswaran, R.; Jagadeesh, S.; Tretiakova, M.S.; Nallasura, V.; Fox, E.A.; Hansen, M.; Schaefer, E.; Naoki, K.; Lader, A.; et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005, 65, 1479–1488. [Google Scholar] [CrossRef]
- Lihavainen, E.; Makela, J.; Spelbrink, J.N.; Ribeiro, A.S. Mytoe: Automatic analysis of mitochondrial dynamics. Bioinformatics 2012, 28, 1050–1051. [Google Scholar] [CrossRef] [PubMed]
- Bouguet, J.-Y. Pyramidal Implementation of the Lucas Kanade Feature Tracker: Description of the Algorithm; Intel Corporation: Santa Clara, CA, USA, 2000. [Google Scholar]
- Gerencser, A.A.; Nicholls, D.G. Measurement of instantaneous velocity vectors of organelle transport: Mitochondrial transport and bioenergetics in hippocampal neurons. Biophys. J. 2008, 95, 3079–3099. [Google Scholar] [CrossRef] [PubMed]
- Karperien, A. FracLac for ImageJ 1999–2013. [CrossRef]
- Waliszewski, P. The Quantitative Criteria Based on the Fractal Dimensions, Entropy, and Lacunarity for the Spatial Distribution of Cancer Cell Nuclei Enable Identification of Low or High Aggressive Prostate Carcinomas. Front. Physiol. 2016, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Halsey, T.C.; Jensen, M.H.; Kadanoff, L.P.; Procaccia, I.I.; Shraiman, B.I. Fractal measures and their singularities: The characterization of strange sets. Phys. Rev. A Gen. Phys. 1986, 33, 1141–1151. [Google Scholar] [CrossRef]
- Lennon, F.E.; Cianci, G.C.; Kanteti, R.; Riehm, J.J.; Arif, Q.; Poroyko, V.A.; Lupovitch, E.; Vigneswaran, W.; Husain, A.; Chen, P.; et al. Unique fractal evaluation and therapeutic implications of mitochondrial morphology in malignant mesothelioma. Sci. Rep. 2016, 6, 24578. [Google Scholar] [CrossRef]
- Havlin, S.; Buldyrev, S.V.; Goldberger, A.L.; Mantegna, R.N.; Ossadnik, S.M.; Peng, C.K.; Simons, M.; Stanley, H.E. Fractals in biology and medicine. Chaos Solitons Fractals 1995, 6, 171–201. [Google Scholar] [CrossRef]
- Mandelbrot, B.B. A Fractal’s Lacunarity, and how it can be Tuned and Measured. In Fractals in Biology and Medicine; Nonnenmacher, T.F., Losa, G.A., Weibel, E.R., Eds.; Birkhäuser Basel: Basel, Switzerland, 1994; pp. 8–21. [Google Scholar]
- Reljin, I.S.; Reljin, B.D. Fractal geometry and multifractals in analyzing and processing medical data and images. Arch. Oncol. 2002, 10, 283–293. [Google Scholar] [CrossRef]
- Lennon, F.E.; Cianci, G.C.; Cipriani, N.A.; Hensing, T.A.; Zhang, H.J.; Chen, C.T.; Murgu, S.D.; Vokes, E.E.; Vannier, M.W.; Salgia, R. Lung cancer-a fractal viewpoint. Nat. Rev. Clin. Oncol. 2015, 12, 664–675. [Google Scholar] [CrossRef]
- Cataldo, A.M.; McPhie, D.L.; Lange, N.T.; Punzell, S.; Elmiligy, S.; Ye, N.Z.; Froimowitz, M.P.; Hassinger, L.C.; Menesale, E.B.; Sargent, L.W.; et al. Abnormalities in mitochondrial structure in cells from patients with bipolar disorder. Am. J. Pathol. 2010, 177, 575–585. [Google Scholar] [CrossRef]
- Calkins, M.J.; Manczak, M.; Mao, P.; Shirendeb, U.; Reddy, P.H. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum. Mol. Genet. 2011, 20, 4515–4529. [Google Scholar] [CrossRef]
- Graves, J.A.; Wang, Y.; Sims-Lucas, S.; Cherok, E.; Rothermund, K.; Branca, M.F.; Elster, J.; Beer-Stolz, D.; Van Houten, B.; Vockley, J.; et al. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS ONE 2012, 7, e37699. [Google Scholar] [CrossRef] [PubMed]
- Campello, S.; Lacalle, R.A.; Bettella, M.; Manes, S.; Scorrano, L.; Viola, A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 2006, 203, 2879–2886. [Google Scholar] [CrossRef] [PubMed]
- Horn, B.K.P.; Schunck, B.G. Determining optical flow. Artif. Intell. 1981, 17, 185–203. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.E.; Liu, X.A.; Puthanveettil, S.V. Automated measurement of fast mitochondrial transport in neurons. Front. Cell. Neurosci. 2015, 9, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, D.J.; Vadakkan, T.J.; Poche, R.A.; Dickinson, M.E. Multifractal and lacunarity analysis of microvascular morphology and remodeling. Microcirculation 2011, 18, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.L.; Nogueira, R.A. Fractal, multifractal and lacunarity analysis applied in retinal regions of diabetic patients with and without nonproliferative diabetic retinopathy. Fractal Geom. Nonlinear Anal. Med. Biol. 2015, 1, 112–119. [Google Scholar]
- Milazzo, L.; Pajor, R. Multifractal analysis of three-dimensional grayscale images: Characterization of natural porous structures. arXiv 2016, arXiv:1605.06009. [Google Scholar]
- Frost, J.J.; Pienta, K.J.; Coffey, D.S. Symmetry and symmetry breaking in cancer: A foundational approach to the cancer problem. Oncotarget 2018, 9, 11429–11440. [Google Scholar] [CrossRef]
- Cagalinec, M.; Safiulina, D.; Liiv, M.; Liiv, J.; Choubey, V.; Wareski, P.; Veksler, V.; Kaasik, A. Principles of the mitochondrial fusion and fission cycle in neurons. J. Cell Sci. 2013, 126 Pt 10, 2187–2197. [Google Scholar] [CrossRef]
- Xu, W.; Foster, B.A.; Richards, M.; Bondioli, K.R.; Shah, G.; Green, C.C. Characterization of prostate cancer cell progression in zebrafish xenograft model. Int. J. Oncol. 2018, 52, 252–260. [Google Scholar] [CrossRef]



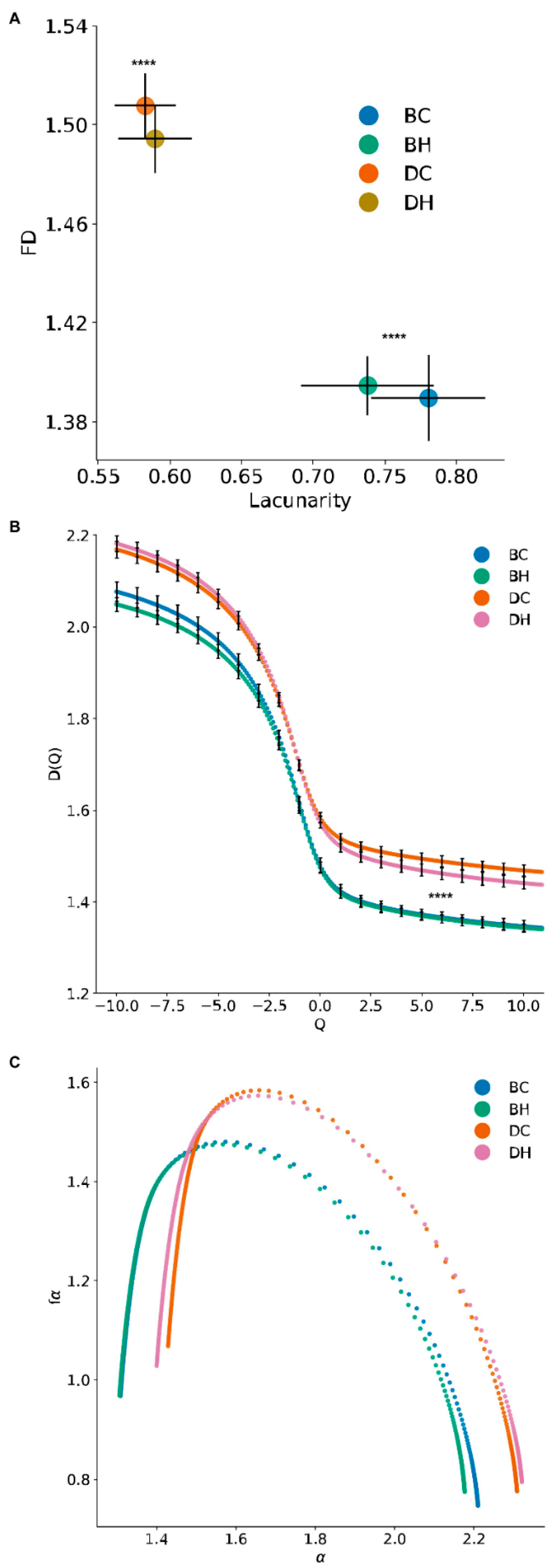

| Normal Cell | SCLC | p-Value (SCLC vs. Normal) | Normal Cell (HGF) | SCLC (HGF) | p-Value (HGF SCLC vs. HGF Normal) | |
|---|---|---|---|---|---|---|
| D-10 | 2.077025 | 2.169330 | 4.8 × 10−4 | 2.049255 | 2.182030 | 4.7 × 10−6 |
| D0 | 1.480179 | 1.584096 | 1.1 × 10−5 | 1.475005 | 1.573209 | 3.5 × 10−6 |
| D1 | 1.423104 | 1.538348 | 9.3 × 10−7 | 1.419077 | 1.521315 | 1.5 × 10−6 |
| D2 | 1.401221 | 1.519859 | 6.9 × 10−7 | 1.398245 | 1.499473 | 9.8 × 10−7 |
| D10 | 1.346371 | 1.468496 | 3.2 × 10−7 | 1.343395 | 1.440852 | 3.2 × 10−6 |
| SCLC | SCLC with Drug | p-Value (SCLC with Drug vs. SCLC) | SCLC (HGF) | SCLC with Drug (HGF) | p-Value (HGF SCLC with Drug vs. HGF SCLC) | |
|---|---|---|---|---|---|---|
| D-10 | 2.169330 | 2.151650 | 0.58 | 2.182030 | 2.129859 | 0.04 |
| D0 | 1.584096 | 1.581537 | 0.60 | 1.573209 | 1.558646 | 0.22 |
| D1 | 1.538348 | 1.526861 | 0.25 | 1.521315 | 1.505344 | 0.15 |
| D2 | 1.519859 | 1.503207 | 0.15 | 1.499473 | 1.482131 | 0.10 |
| D10 | 1.468496 | 1.446126 | 0.11 | 1.440852 | 1.422356 | 0.08 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirzapoiazova, T.; Li, H.; Nathan, A.; Srivstava, S.; Nasser, M.W.; Lennon, F.; Armstrong, B.; Mambetsariev, I.; Chu, P.G.; Achuthan, S.; et al. Monitoring and Determining Mitochondrial Network Parameters in Live Lung Cancer Cells. J. Clin. Med. 2019, 8, 1723. https://doi.org/10.3390/jcm8101723
Mirzapoiazova T, Li H, Nathan A, Srivstava S, Nasser MW, Lennon F, Armstrong B, Mambetsariev I, Chu PG, Achuthan S, et al. Monitoring and Determining Mitochondrial Network Parameters in Live Lung Cancer Cells. Journal of Clinical Medicine. 2019; 8(10):1723. https://doi.org/10.3390/jcm8101723
Chicago/Turabian StyleMirzapoiazova, Tamara, Haiqing Li, Anusha Nathan, Saumya Srivstava, Mohd W. Nasser, Frances Lennon, Brian Armstrong, Isa Mambetsariev, Peiguo G. Chu, Srisairam Achuthan, and et al. 2019. "Monitoring and Determining Mitochondrial Network Parameters in Live Lung Cancer Cells" Journal of Clinical Medicine 8, no. 10: 1723. https://doi.org/10.3390/jcm8101723








