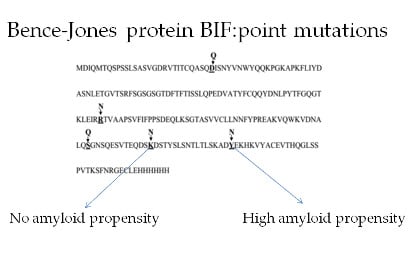
Effect of Single Amino Acid Substitutions by Asn and Gln on Aggregation Properties of Bence-Jones Protein BIF
Abstract
:1. Introduction
2. Results
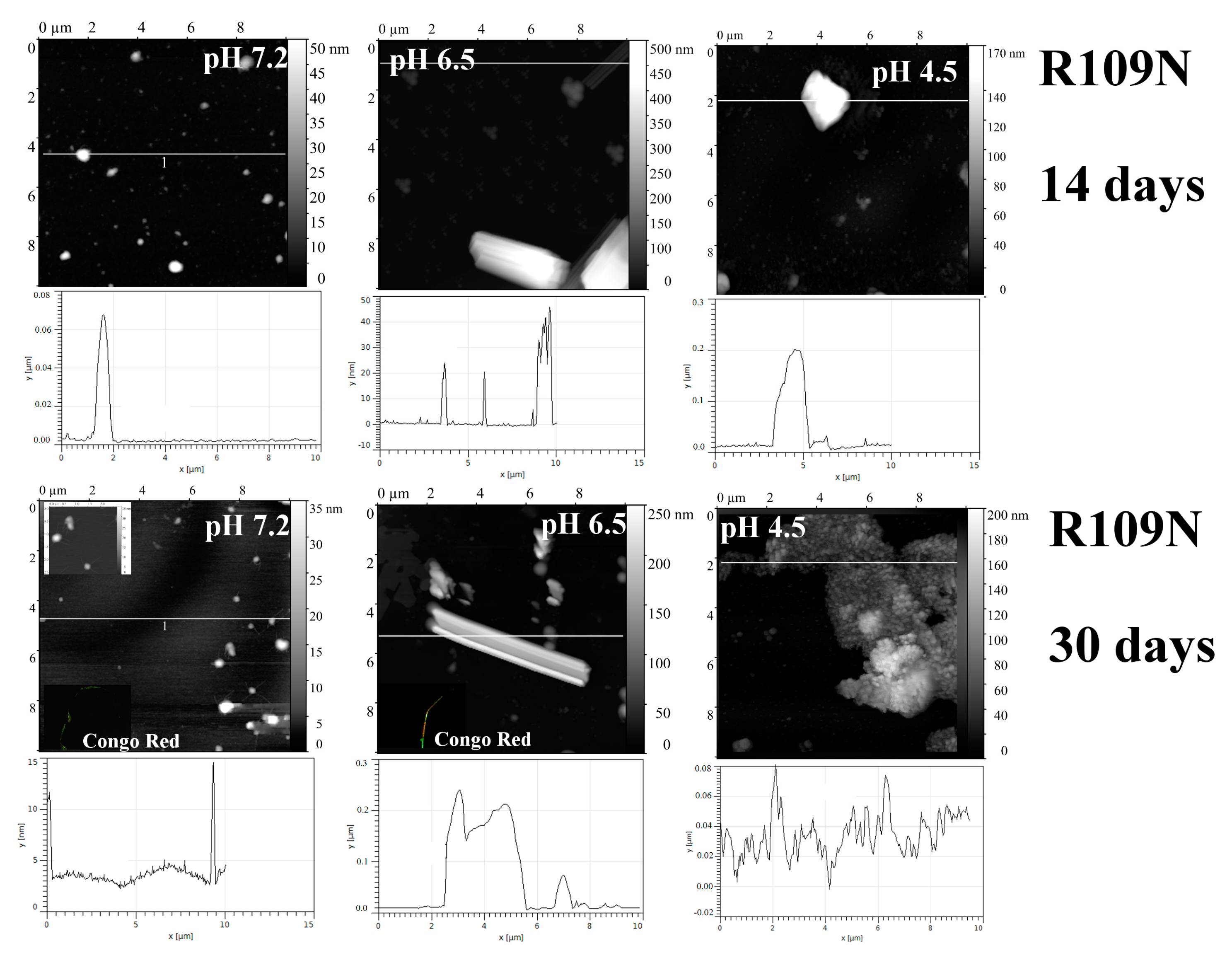
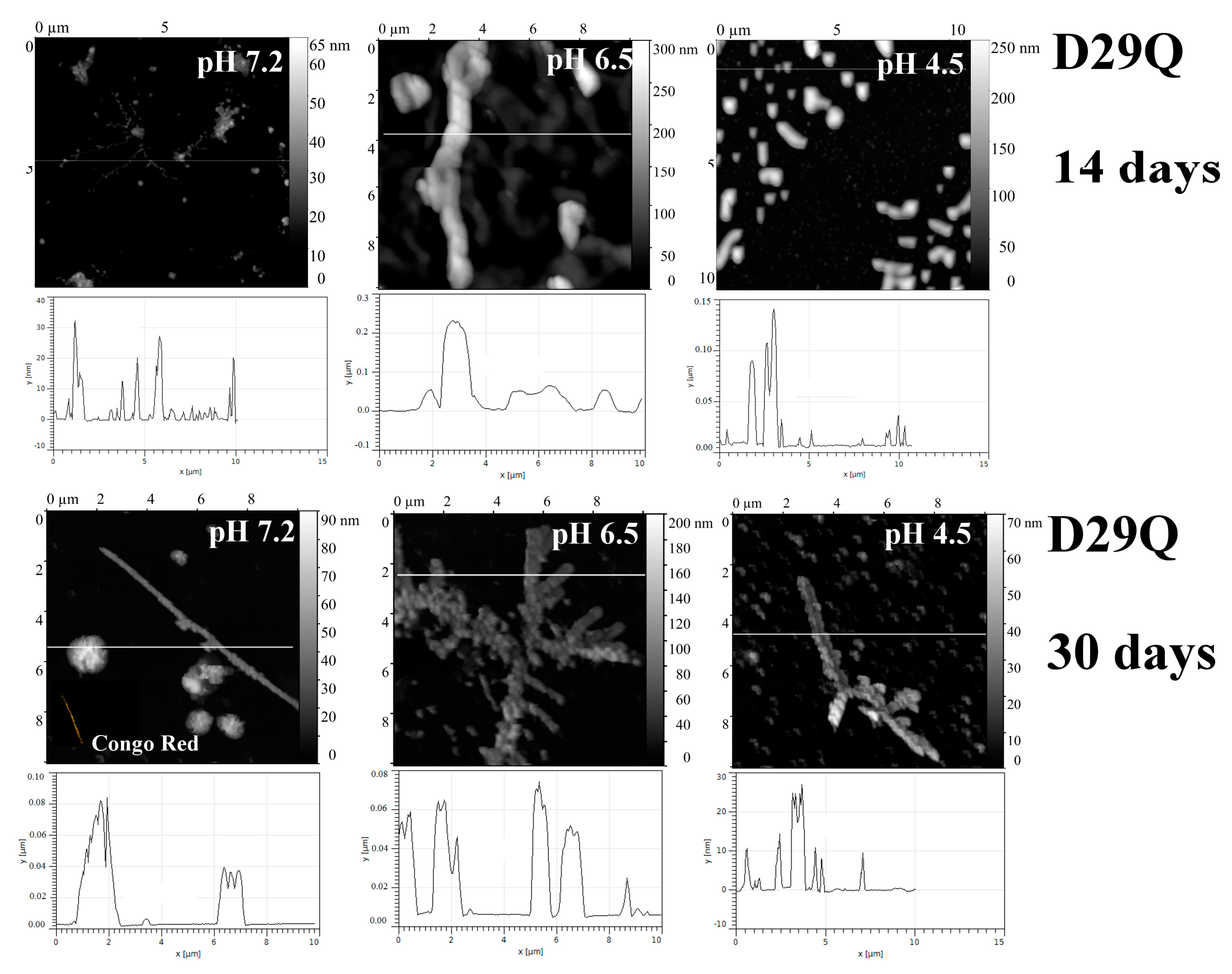
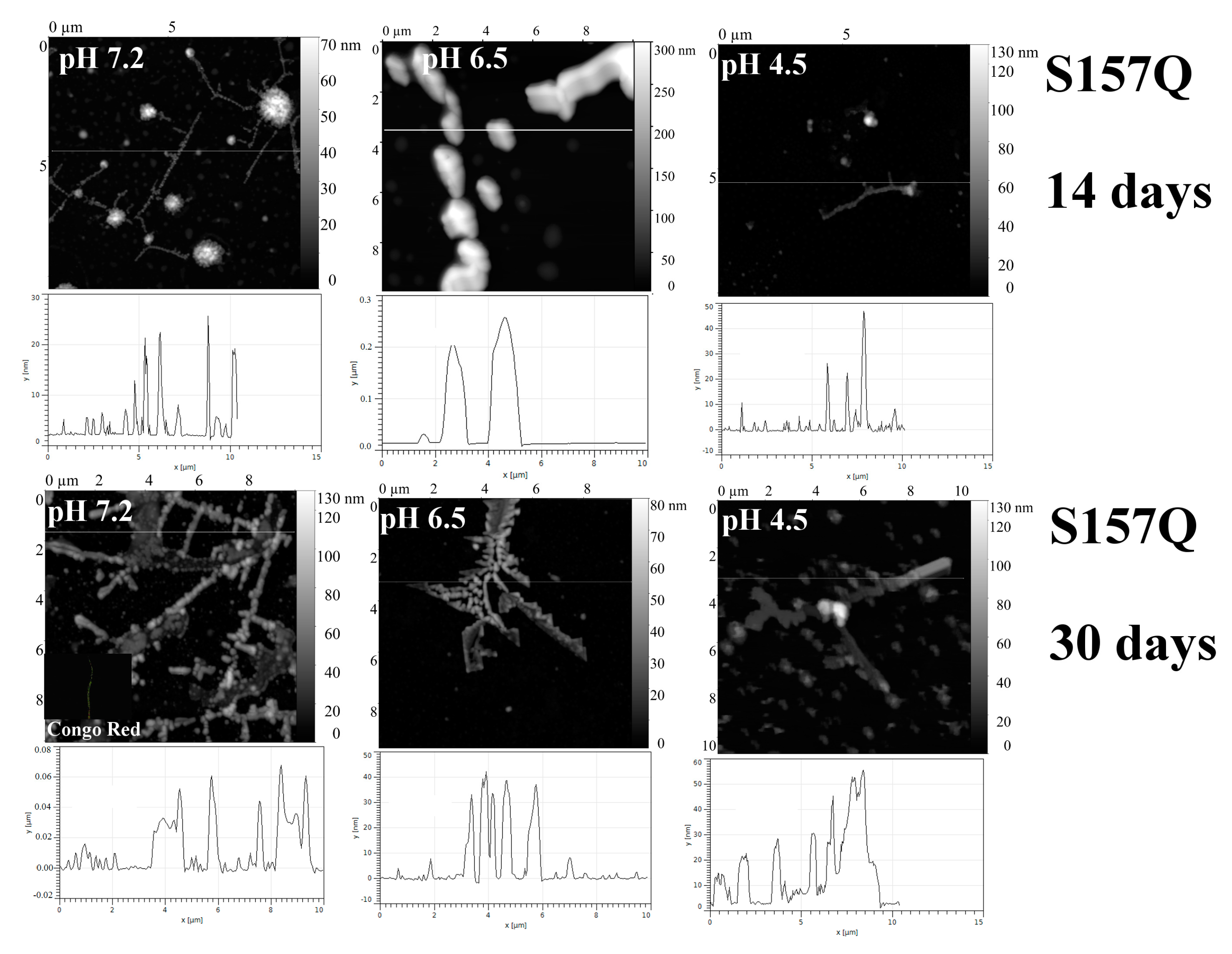
2.1. Analysis of Morphology of Aggregates
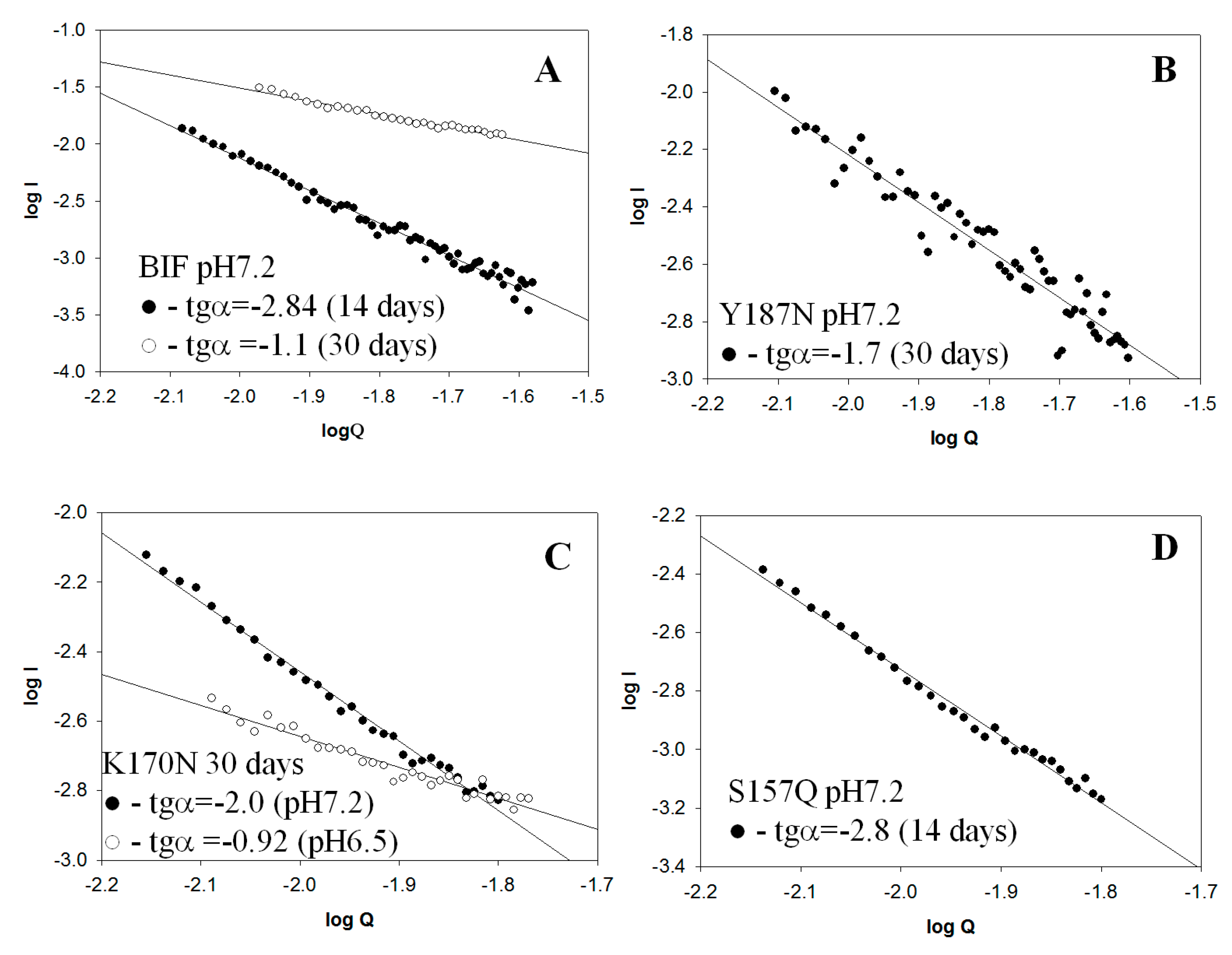
2.2. Small Angle X-Ray Scattering (SAXS) Data
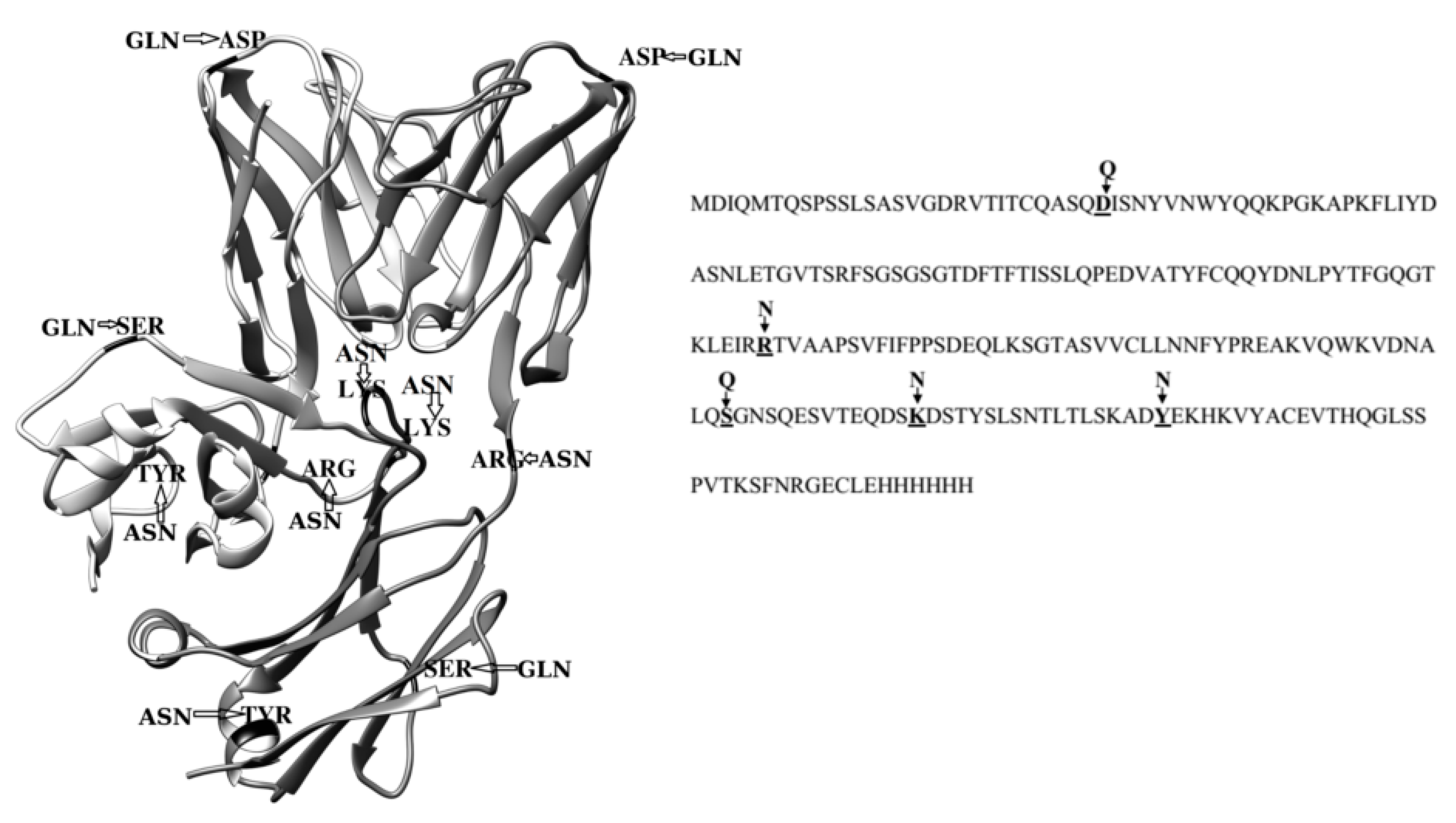
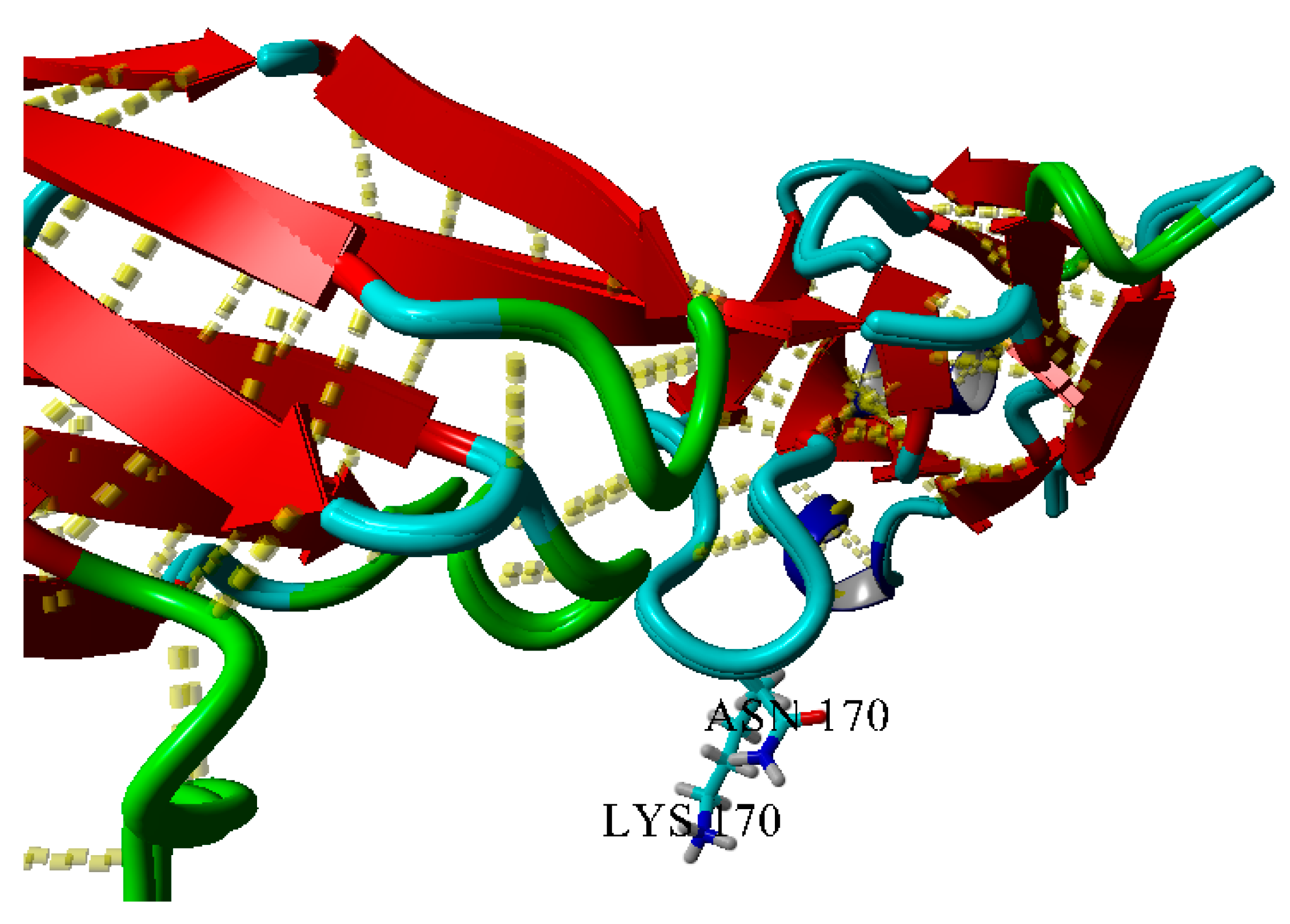
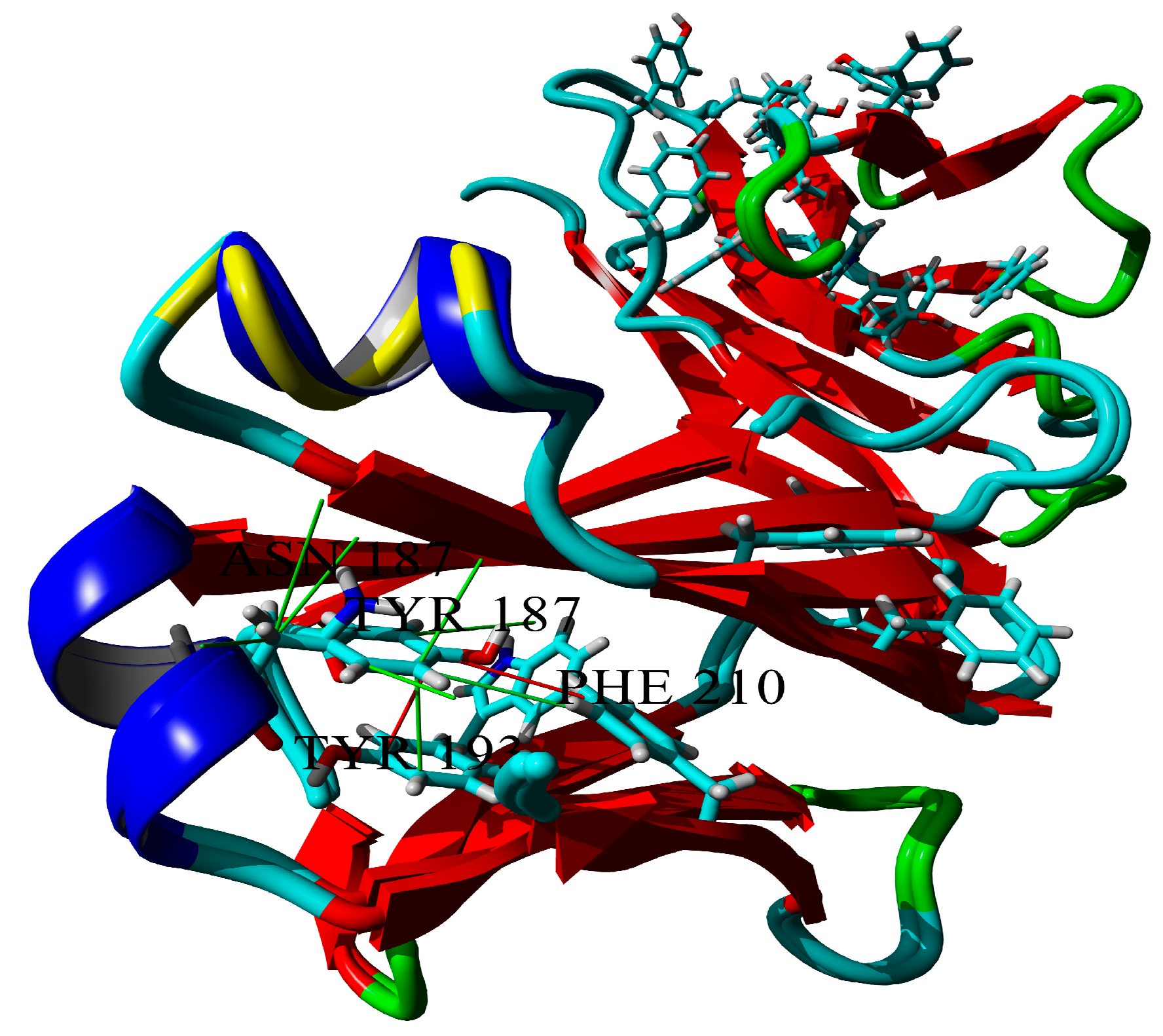
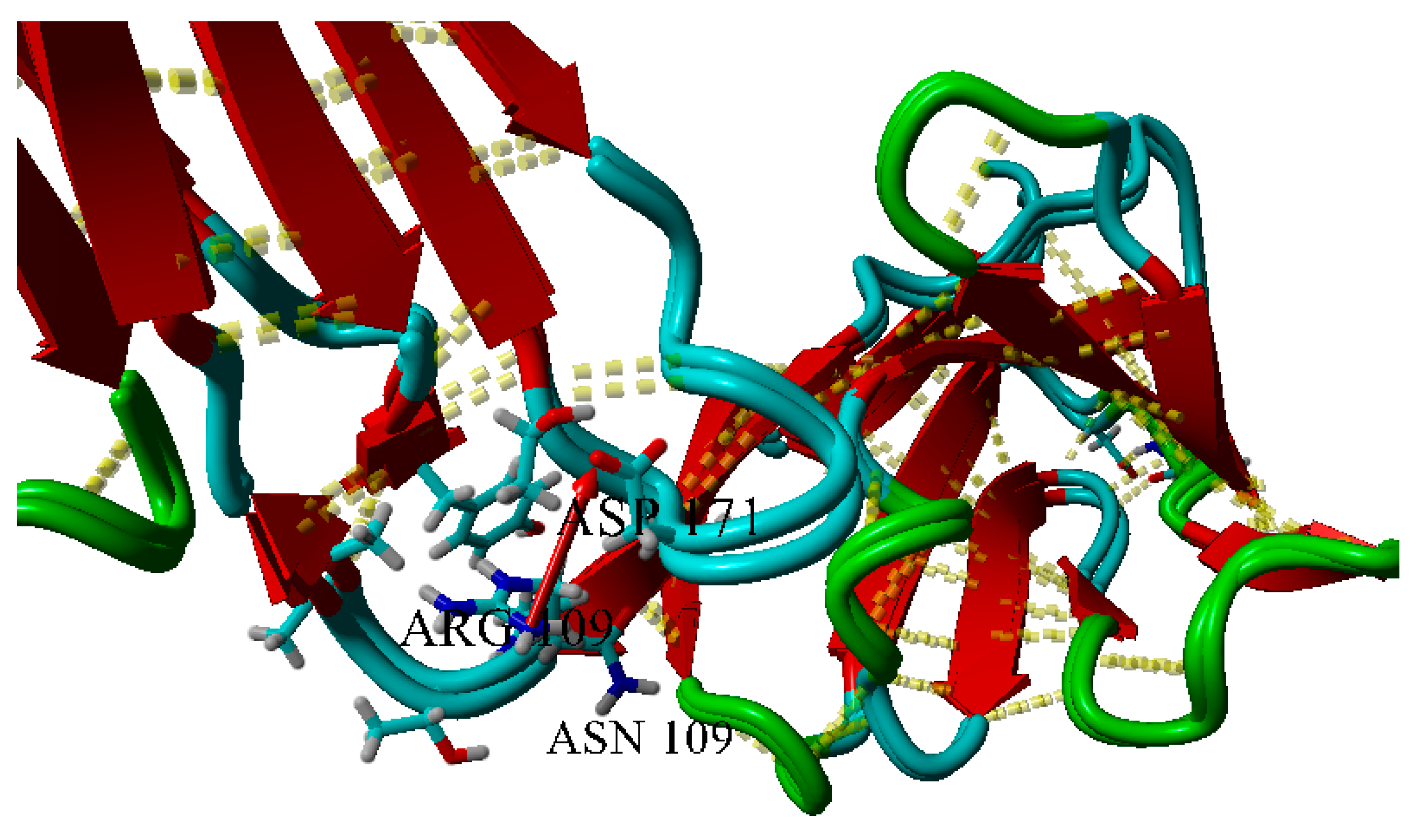
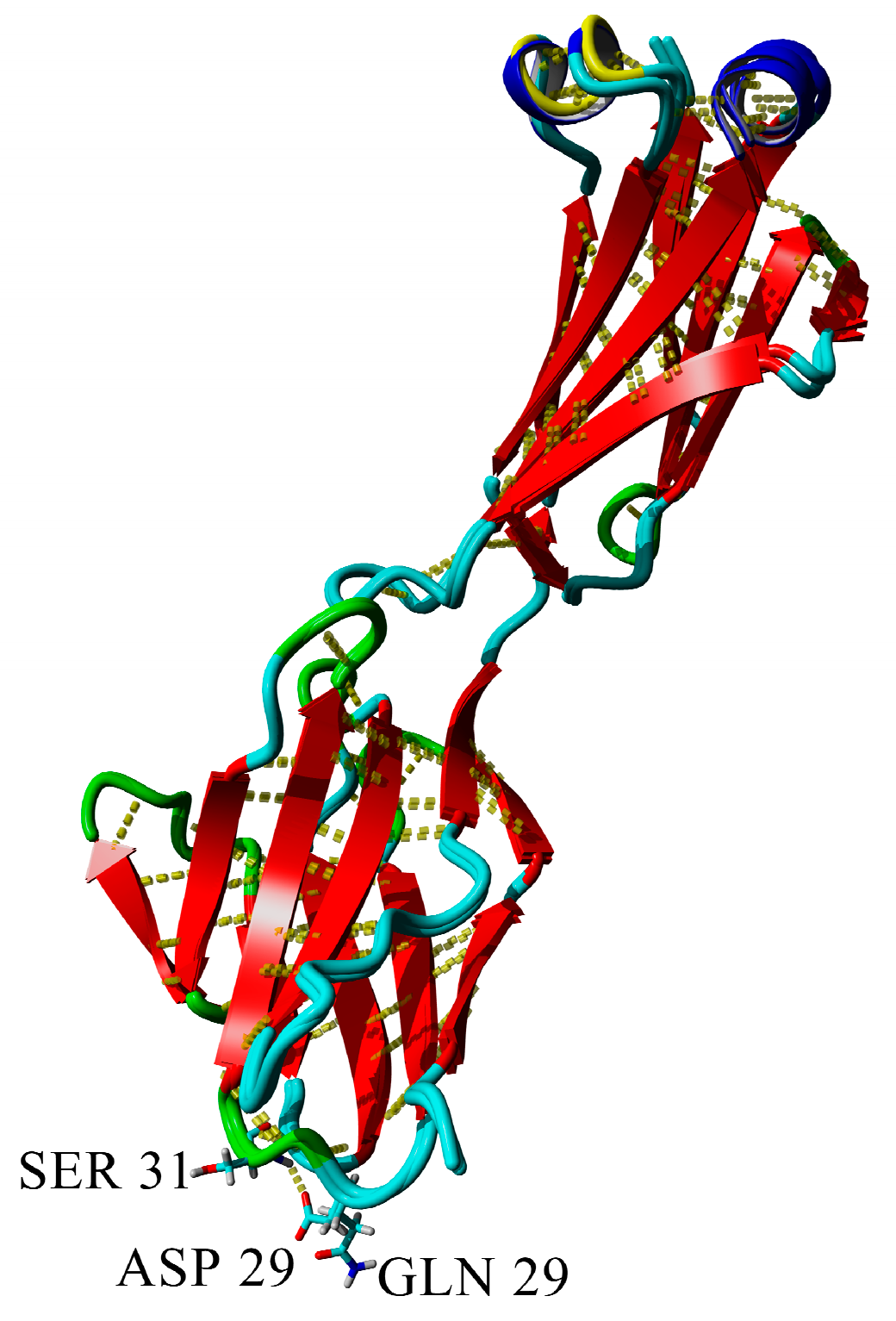
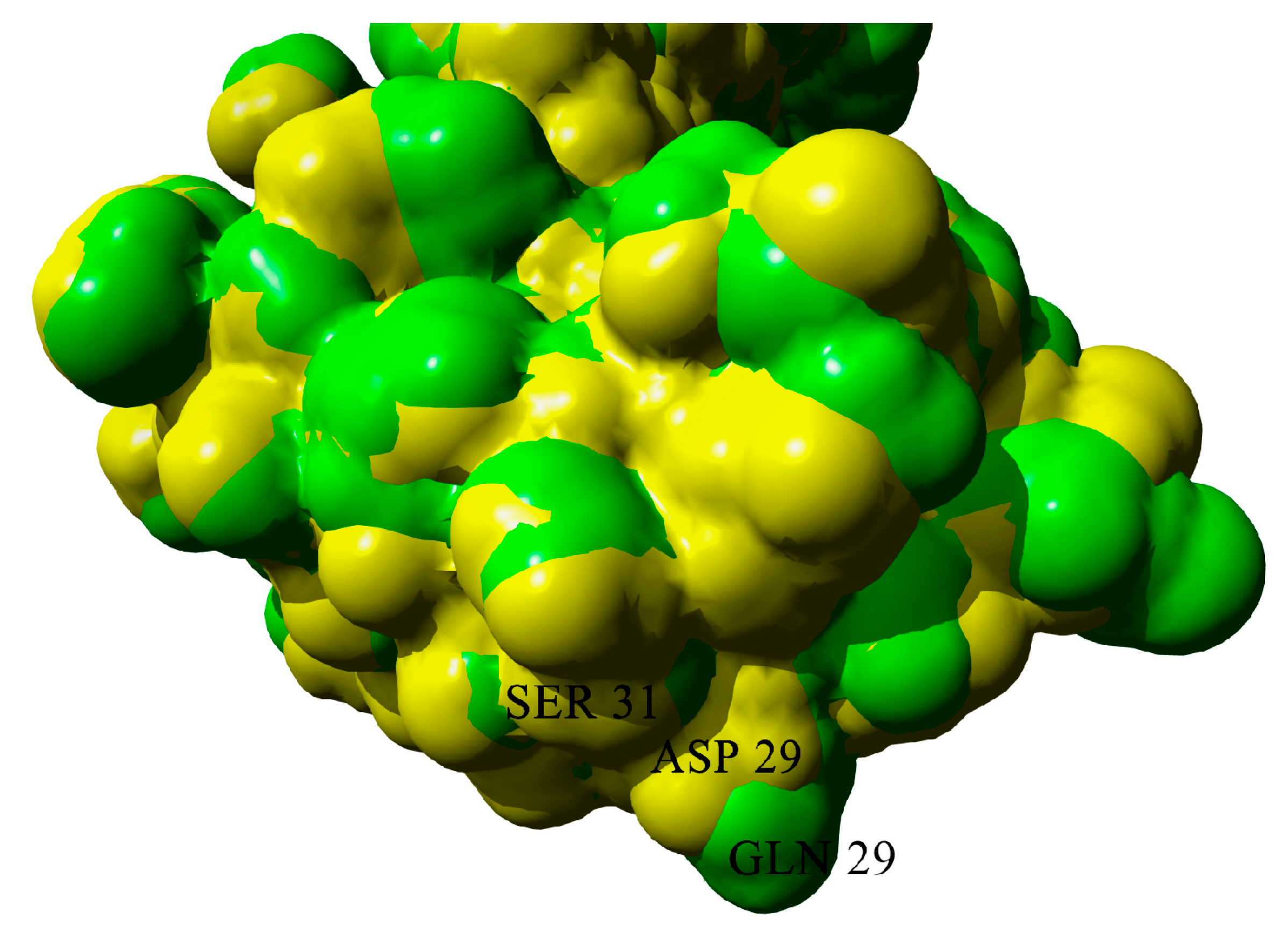
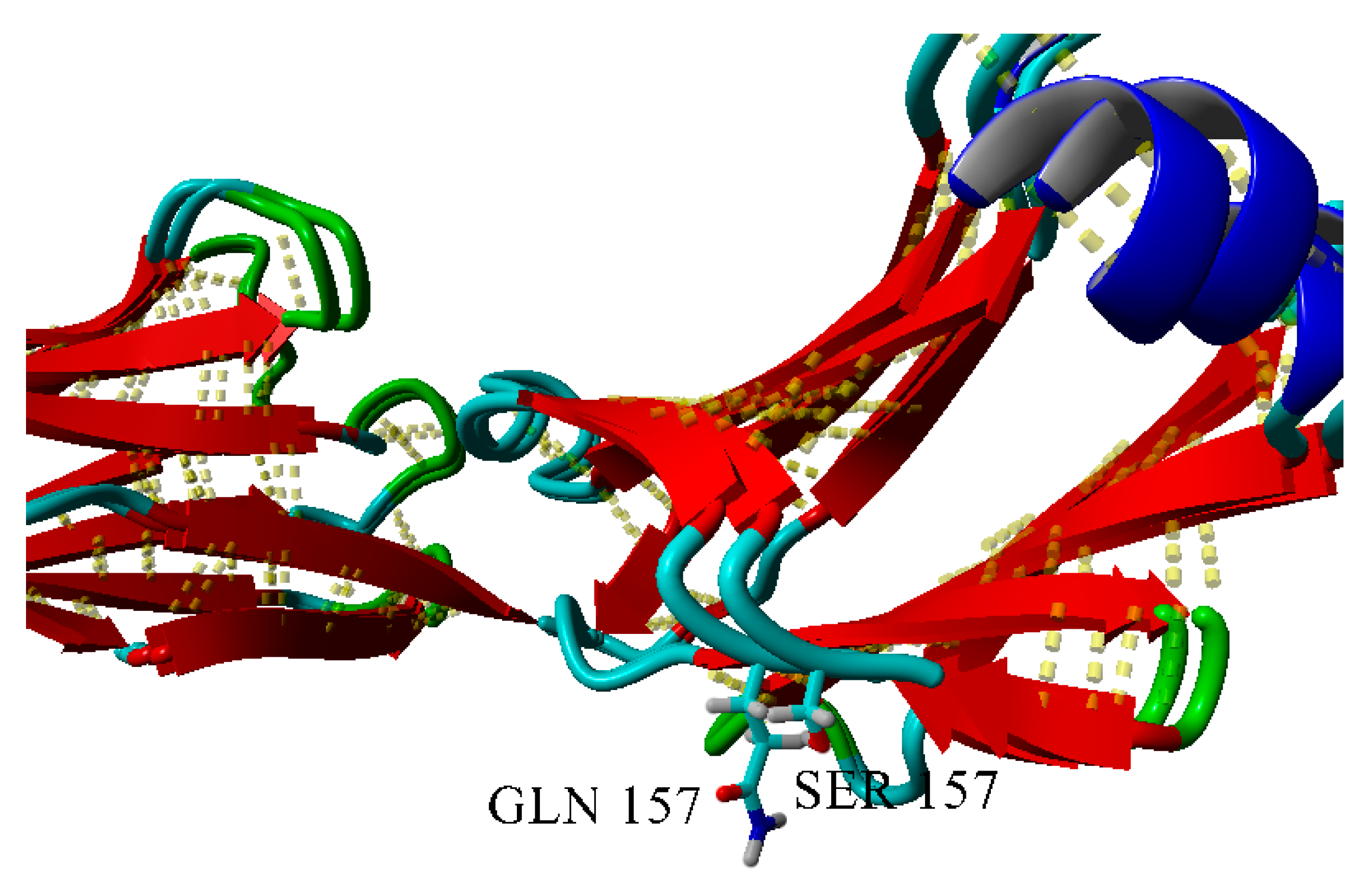
2.3. Dimerization of BIF Molecules: Role of Surface Residue Substitutions
3. Discussion
4. Materials and Methods
4.1. Construction of the BIF-Expressing Plasmid pETBIF
4.2. Site-Directed Mutagenesis
4.3. Expression and Purification of the Recombinant BIF Protein and Its Mutant Analogues
4.4. Refolding of the Recombinant BIF Protein and Its Mutant Analogues
4.5. Formation of Aggregates of BIF Protein and Its Mutant Analogues in Solution
4.6. The Thioflavin T (ThT) Assay
4.7. Congo Red Spectroscopic Assay
4.8. Electrophoretic Analysis of Aggregates in Polyacrylamide Gel (PAGE) with 0.1% Sodium Dodecyl Sulfate
4.9. AFM Measurements
4.10. Small Angle X-Ray Scattering (SAXS)
4.11. The Congo Red Birefringence Assay
4.12. Molecular Modelling
4.13. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Protein | pH | tgα (SAXS), Type of Aggregates | Binding of ThT * (30 Days) | Binding of CR * (30 Days) | Resistance to 2% SDS (30 Days) | Birefringence of CR (30 Days) |
|---|---|---|---|---|---|---|
| BIF | 7.2 | −2.84 Mainly branched flat associates (14 days) −1.1 Mainly fibrillar structures (30 days) | + | + | + | + |
| 6.5 | “—” * | + | + | + | + | |
| 4.5 | −3.42 Mainly branched structures from globular particles | − | − | − | − | |
| K170N | 7.2 | −2.0 Mainly branched flat associates (30 days) | − | − | − | − |
| 6.5 | −0.92 Rod-like structures (30 days) | − | − | − | − | |
| 4.5 | “—” | − | − | − | − | |
| Y187N | 7.2 | −1.7 Mainly rod-like structures (30 days) | + | + | + | + |
| 6.5 | “—” | + | + | + | + | |
| 4.5 | “—” | + | + | + | + | |
| R109N | 7.2 | “—” | + | + | + | + |
| 6.5 | “—” | + | + | + | + | |
| 4.5 | “—” | − | − | − | − | |
| S157Q | 7.2 | −2.8 Mainly branched flat associates (14 days) | + | + | + | + |
| 6.5 | “—” | − | − | − | − | |
| 4.5 | “—” | − | − | − | − | |
| D29Q | 7.2 | “—” | + | + | + | + |
| 6.5 | “—” | − | − | − | − | |
| 4.5 | “—” | − | − | − | − |
References
- Solomon, A.; Weiss, D.T.; Murphy, C.L.; Hrncic, R.; Wall, J.S.; Schell, M. Light chain-associated amyloid deposits comprised of a novel kappa constant domain. Proc. Natl. Acad. Sci. USA 1998, 95, 9547–9551. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, H.; Zhu, L.; Zhou, J.M.; Perrett, S. Amyloid nucleation and hierarchical assembly of Ure2p fibrils. Role of asparagine/glutamine repeat and nonrepeat regions of the prion domains. J. Biol. Chem. 2004, 279, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.F. Glutamine repeats and neurodegenerative diseases: Molecular aspects. Trends Biol. Sci. 1999, 24, 58–63. [Google Scholar] [CrossRef]
- Haass, C.; Steiner, H. Protofibrils, the unifying toxic molecule of neurodegenerative disorders? Nat. Neurosci. 2001, 4, 859–860. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.M.; Klyubin, I.; Fadeeva, J.V.; Cullen, W.K.; Anwyl, R.; Wolfe, M.S.; Rowan, M.J.; Selkoe, D.J. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002, 416, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Timchenko, M.A.; Timchenko, A.A. Influence of a single point mutation in the constant domain of the Bence-Jones Protein BIF on its aggregation properties. Biochem. (Mosc.) 2018, 83, 107–118. [Google Scholar] [CrossRef]
- Sabate, R.; Rousseau, F.; Schymkowitz, J.; Batlle, C.; Ventura, S. Amyloids or prions? That is the question. Prion 2015, 9, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Nokwe, C.N.; Hora, M.; Zacharias, M.; Yagi, H.; John, C.; Reif, B.; Goto, Y.; Buchner, J. The Antibody Light-Chain Linker Is Important for Domain Stability and Amyloid Formation. J. Mol. Biol. 2015, 427, 3572–3586. [Google Scholar] [CrossRef]
- Weber, B.; Hora, M.; Kazman, P.; Göbl, C.; Camilloni, C.; Reif, B.; Buchner, J. The Antibody Light-Chain Linker Regulates Domain Orientation and Amyloidogenicity. J. Mol. Biol. 2018, 430, 4925–4940. [Google Scholar] [CrossRef] [Green Version]
- Roussel, A.; Spinelli, S.; Déret, S.; Navaza, J.; Aucouturier, P.; Cambillau, C. The structure of an entire noncovalent immunoglobulin kappa light-chain dimer (Bence-Jones protein) reveals a weak and unusual constant domains association. Eur. J. Biochem. 1999, 260, 192–199. [Google Scholar] [CrossRef]
- Kabat, E.A.; Wu, T.T.; Perry, H.M.; Gottesman, K.S.; Foeller, C. Sequences of Proteins of Immunologic Interest. Nat. Inst. Health 1991, 1, 91–32421. Available online: https://books.google.ru/books?id=3jMvZYW2ZtwC&printsec=frontcover&dq=Sequences+of+Proteins+of+Immunologic+Interest.&hl=en&sa=X&ved=0ahUKEwiv4quV6qjlAhVOwMQBHcauC6cQ6AEIKzAA#v=onepage&q=Sequences%20of%20Proteins%20of%20Immunologic%20Interest.&f=false (accessed on 19 October 2019).
- Myatt, E.A.; Westholm, F.A.; Weiss, D.T.; Solomon, A.; Schiffer, M.; Stevens, F.J. Pathogenic potential of human monoclonal immunoglobulin light chains: Relationship of in vitro aggregation to in vivo organ deposition. Proc. Natl. Acad. Sci. USA 1994, 91, 3034–3038. [Google Scholar] [CrossRef] [PubMed]
- Katina, N.; Timchenko, A.; Balobanov, V.; Vasiliev, V.; Kashparov, I.; Bychkova, V.; Kihara, H. Kinetics of mutant apomyoglobin association. Macromol. Symp. 2012, 317, 215–226. [Google Scholar] [CrossRef]
- Feigin, L.A.; Svergun, D.A. Structure Analysis by Small-Angle X-Ray and Neutron Scattering; Plenum Press: New York, NY, USA, 1987. [Google Scholar]
- Helms, L.R.; Wetzel, R. Destabilizing loop swaps in the CDRs of an immunoglobulin VL domain. Protein Sci. 1995, 4, 2073–2081. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.; Schell, M.; Murphy, C.; Hrncic, R.; Stevens, F.J.; Solomon, A. Thermodynamic instability of human lambda 6 light chains: Correlation with fibrillogenicity. Biochemistry 1999, 38, 14101–14108. [Google Scholar] [CrossRef] [PubMed]
- Raffen, R.; Diekman, L.J.; Szpunar, M.; Wanschl, C.; Rokkuludi, P.R.; Dave, R. Physicochemical consequences of amino acid variations that contribute to fibril formation by immunoglobulin light chains. Protein Sci. 1999, 8, 509–517. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Wall, J.S.; Meyer, J.; Murphy, C.; Randolph, T.W.; Manning, M. Thermodynamic modulation of light chain amyloid fibril formation. J. Biol. Chem. 2000, 275, 1570–1574. [Google Scholar] [CrossRef]
- Bliznyukov, O.P.; Kozmin, L.D.; Vysotskaya, L.L.; Golenkov, A.K.; Tishchenko, V.M.; Samoylovich, M.P.; Klimovich, V.B. Human immunoglobulin light chains lambda form amyloid fibrils and granular aggregates in solution. Biochemistry 2005, 70, 458–466. [Google Scholar]
- Stevens, F.J.; Myatt, E.A.; Chang, C.H.; Westholm, F.A.; Eulitz, M.; Weiss, D.T.; Murphy, C.; Solomon, A.; Schiffer, M. A molecular model for self-assembly of amyloid fibrils: Immunoglobulin light chains. Biochemistry 1995, 34, 10697–10702. [Google Scholar] [CrossRef]
- Nowak, M. Immunoglobulin kappa light chain and its amyloidogenic mutants: A molecular dynamics study. Proteins 2004, 55, 11–21. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pondaven, S.P.; Jaroniec, C.P. Conformational flexibility of a human immunoglobulin light chain variable domain by relaxation dispersion nuclear magnetic resonance spectroscopy: Implications for protein misfolding and amyloid assembly. Biochemistry 2011, 50, 5845–5857. [Google Scholar] [CrossRef] [PubMed]
- Wilkins-Stevens, P.; Raffen, R.; Hanson, D.K.; Deng, Y.L.; Berrios-Hammond, M.; Westholm, F.A.; Murphy, C.; Eulitz, M.; Wetzel, R.; Solomon, A.; et al. Recombinant immunoglobulin variable domains generated from synthetic genes provide a system for in vitro characterization of light-chain amyloid proteins. Protein Sci. 1995, 4, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Maniatis, T.; Fritisch, E.F.; Sambrok, J. A Laboratory Manual. In Molecular Cloning; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1982; p. 545. [Google Scholar]
- LaemmLi, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Dubnovitsky, A.P.; Kravchuk, Z.I.; Chumanevich, A.A.; Cozzi, A.; Arosio, P.; Martsev, S.P. Expression, refolding, and ferritin-binding activity of the isolated VL-domain of monoclonal antibody F11. Biochemistry 2000, 65, 1011–1018. [Google Scholar] [PubMed]
- LeVine, H., 3rd. Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef]
- Klunk, W.E.; Jacob, R.F.; Mason, R.P. Quantifying amyloid beta-peptide (Abeta) aggregation using the Congo Red-Abeta (CR-abeta) spectrophotometric assay. Anal. Biochem. 1999, 266, 66–76. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Alexandrov, I.M.; Mitkevich, O.V.; Shkundina, I.S.; Ter-Avanesyan, M.D. Purification and analysis of prion and amyloid aggregates. Methods 2006, 39, 50–55. [Google Scholar] [CrossRef]
- Glatter, O.; Kratky, O. Small Angle X-Ray Scattering; Academic Press: Cambridge, MA, USA, 1982; p. 515. [Google Scholar]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 11, 2507–2524. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timchenko, M.; Abdullatypov, A.; Kihara, H.; Timchenko, A. Effect of Single Amino Acid Substitutions by Asn and Gln on Aggregation Properties of Bence-Jones Protein BIF. Int. J. Mol. Sci. 2019, 20, 5197. https://doi.org/10.3390/ijms20205197
Timchenko M, Abdullatypov A, Kihara H, Timchenko A. Effect of Single Amino Acid Substitutions by Asn and Gln on Aggregation Properties of Bence-Jones Protein BIF. International Journal of Molecular Sciences. 2019; 20(20):5197. https://doi.org/10.3390/ijms20205197
Chicago/Turabian StyleTimchenko, Maria, Azat Abdullatypov, Hiroshi Kihara, and Alexander Timchenko. 2019. "Effect of Single Amino Acid Substitutions by Asn and Gln on Aggregation Properties of Bence-Jones Protein BIF" International Journal of Molecular Sciences 20, no. 20: 5197. https://doi.org/10.3390/ijms20205197