MicroRNA Profiling During Neural Differentiation of Induced Pluripotent Stem Cells
Abstract
:1. Introduction
2. Results
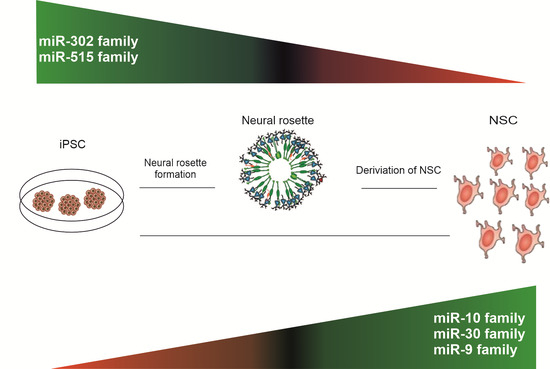
2.1. Derivation of NSC from Human iPSC
2.2. miRNA Expression Profiling during Neural Differentiation from Induced Pluripotent Stem Cells
2.3. Biological Processes Regulated by miRNA During Neural Differentiation
3. Discussion
4. Materials and Methods
4.1. Neural Induction of iPSC
4.2. Total RNA Isolation and RT-qPCR
4.3. RT-qPCR for miRNA Expression
4.4. Immunocytochemistry
4.5. Microarray
4.6. miRNA-Target Gene Prediction
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ECM | Extracellular matrix |
| KLF4 | Kruppel-like factor 4 |
| WNT10B | Wingless-type MMTV integration site family, member 10B |
| BMP | Bone morphogenetic protein |
| c-Myc | MYC proto-oncogene |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
| EDTA | Ethylene diamine tetra acetic |
| ESC | Embryonic stem cells |
| FZD4/5/8 | Frizzled4/5/8 |
| GEO | Gene expression omnibus |
| GO | Gene ontology |
| GPCCi001-A | Human induced pluripotent stem cell line created in Greater Poland Cancer Centre |
| iPSC | Induced pluripotent stem cells |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NANOG | Nanog homeobox |
| NR2F2 | Nuclear receptor subfamily 2, group F, member 2 |
| NSC | Neuronal stem cells |
| OCT3/4 | Octamer-binding transcription factor 4 |
| PAX6 | Paired box protein 6 |
| PBS | Phosphate buffer saline |
| PCR | Polymerase chain reaction |
| POU5F1 | POU class 5 homeobox 1 |
| RT-qPCR | Real time quantitative polymerase chain reaction |
| SMAD4 | SMAD family member 4 |
| SOX1 | SRY (Sex determining region Y)-box 1 |
| SOX2 | SRY (Sex determining region Y)-box 2 |
| TGF-β | Transforming growth factor beta |
| WNT | Wingless-type MMTV integration site family |
| WNT3A | Wingless-type MMTV integration site family, member 3A |
| WNT4 | Wingless-type MMTV integration site family, member 4 |
| ZEB | Zinc finger E-box binding homeobox |
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Abematsu, M.; Falk, A.; Tsujimura, K.; Sanosaka, T.; Juliandi, B.; Semi, K.; Namihira, M.; Komiya, S.; Smith, A.; et al. Treatment of a mouse model of spinal cord injury by transplantation of human induced pluripotent stem cell-derived long-term self-renewing neuroepithelial-like stem cells. Stem Cells 2012, 30, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, H.; Ikuno, T.; Takeda, M.; Fukushima, H.; Marui, A.; Katayama, S.; Shimizu, T.; Ikeda, T.; Okano, T.; Sakata, R.; et al. Human ips cell-engineered cardiac tissue sheets with cardiomyocytes and vascular cells for cardiac regeneration. Sci. Rep. 2014, 4, 6716. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Takayama, N.; Hirata, S.; Seo, H.; Endo, H.; Ochi, K.; Fujita, K.; Koike, T.; Harimoto, K.; Dohda, T.; et al. Expandable megakaryocyte cell lines enable clinically applicable generation of platelets from human induced pluripotent stem cells. Cell Stem Cell 2014, 14, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Shahjalal, H.M.; Shiraki, N.; Sakano, D.; Kikawa, K.; Ogaki, S.; Baba, H.; Kume, K.; Kume, S. Generation of insulin-producing beta-like cells from human ips cells in a defined and completely xeno-free culture system. J. Mol. Cell Biol. 2014, 6, 394–408. [Google Scholar] [CrossRef]
- Li, Y.; Wu, W.H.; Hsu, C.W.; Nguyen, H.V.; Tsai, Y.T.; Chan, L.; Nagasaki, T.; Maumenee, I.H.; Yannuzzi, L.A.; Hoang, Q.V.; et al. Gene therapy in patient-specific stem cell lines and a preclinical model of retinitis pigmentosa with membrane frizzled-related protein defects. Mol. Ther. 2014, 22, 1688–1697. [Google Scholar] [CrossRef]
- Ma, N.; Liao, B.; Zhang, H.; Wang, L.; Shan, Y.; Xue, Y.; Huang, K.; Chen, S.; Zhou, X.; Chen, Y.; et al. Transcription activator-like effector nuclease (talen)-mediated gene correction in integration-free beta-thalassemia induced pluripotent stem cells. J. Biol. Chem. 2013, 288, 34671–34679. [Google Scholar] [CrossRef]
- Song, B.; Fan, Y.; He, W.; Zhu, D.; Niu, X.; Wang, D.; Ou, Z.; Luo, M.; Sun, X. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by crispr/cas9 system. Stem Cells Dev. 2015, 24, 1053–1065. [Google Scholar] [CrossRef]
- An, M.C.; Zhang, N.; Scott, G.; Montoro, D.; Wittkop, T.; Mooney, S.; Melov, S.; Ellerby, L.M. Genetic correction of huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell 2012, 11, 253–263. [Google Scholar] [CrossRef]
- Xia, G.; Santostefano, K.; Hamazaki, T.; Liu, J.; Subramony, S.H.; Terada, N.; Ashizawa, T. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J. Mol. Neurosci. 2013, 51, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Asai, M.; Tsukita, K.; Kutoku, Y.; Ohsawa, Y.; Sunada, Y.; Imamura, K.; Egawa, N.; Yahata, N.; Okita, K.; et al. Modeling alzheimer’s disease with ipscs reveals stress phenotypes associated with intracellular abeta and differential drug responsiveness. Cell Stem Cell 2013, 12, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, J.; Zhang, L.; Xu, H.; Guo, X.; Deng, S.; Liu, L.; Yu, D.; Chen, Y.; Li, Z. Generation of the scn1a epilepsy mutation in hips cells using the talen technique. Sci. Rep. 2014, 4, 5404. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, K.; Du, Z.; Cao, J.; Petersen, A.; Liu, H.; Blackbourn, L.W.T.; Huang, C.L.; Errigo, A.; Yin, Y.; et al. Modeling als with ipscs reveals that mutant sod1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 2014, 14, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Merkle, F.T.; Alvarez-Buylla, A. Neural stem cells in mammalian development. Curr. Opin. Cell Biol. 2006, 18, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian micrornas uncovers a subset of brain-expressed micrornas with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific micrornas. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific micrornas from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, X.; Hsieh, J.; Wichterle, H.; Impey, S.; Banerjee, S.; Neveu, P.; Kosik, K.S. Microrna regulation of neural stem cells and neurogenesis. J. Neurosci. 2010, 30, 14931–14936. [Google Scholar] [CrossRef]
- Lach, M.S.; Wroblewska, J.P.; Augustyniak, E.; Kulcenty, K.; Suchorska, W.M. A feeder- and xeno-free human induced pluripotent stem cell line obtained from primary human dermal fibroblasts with epigenetic repression of reprogramming factors expression: Gpcci001-a. Stem Cell Res. 2017, 20, 34–37. [Google Scholar] [CrossRef]
- Venkatesh, K.; Kumari, A.; Sen, D. Microrna signature changes during induction of neural stem cells from human mesenchymal stem cells. Nanomedicine 2019, 17, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.A.; Park, H.; Lim, E.H.; Lee, K.W. Microrna expression profiling in neurogenesis of adipose tissue-derived stem cells. J. Genet. 2011, 90, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microrna-9 and nuclear receptor tlx in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Kurokawa, D.; Nakao, H.; Ohmura, T.; Aizawa, S. Microrna-9 modulates cajal-retzius cell differentiation by suppressing foxg1 expression in mouse medial pallium. J. Neurosci. 2008, 28, 10415–10421. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. Mir-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.K. The microrna mir-124 antagonizes the anti-neural rest/scp1 pathway during embryonic cns development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Lipchina, I.; Elkabetz, Y.; Hafner, M.; Sheridan, R.; Mihailovic, A.; Tuschl, T.; Sander, C.; Studer, L.; Betel, D. Genome-wide identification of microrna targets in human es cells reveals a role for mir-302 in modulating bmp response. Genes Dev. 2011, 25, 2173–2186. [Google Scholar] [CrossRef]
- Rosa, A.; Brivanlou, A.H. A regulatory circuitry comprised of mir-302 and the transcription factors oct4 and nr2f2 regulates human embryonic stem cell differentiation. EMBO J. 2011, 30, 237–248. [Google Scholar] [CrossRef]
- Du, Z.W.; Ma, L.X.; Phillips, C.; Zhang, S.C. Mir-200 and mir-96 families repress neural induction from human embryonic stem cells. Development 2013, 140, 2611–2618. [Google Scholar] [CrossRef]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microrna genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar] [CrossRef]
- Houbaviy, H.B.; Dennis, L.; Jaenisch, R.; Sharp, P.A. Characterization of a highly variable eutherian microrna gene. RNA 2005, 11, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Lichner, Z.; Pall, E.; Kerekes, A.; Pallinger, E.; Maraghechi, P.; Bosze, Z.; Gocza, E. The mir-290-295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation 2011, 81, 11–24. [Google Scholar] [CrossRef]
- Stappert, L.; Roese-Koerner, B.; Brustle, O. The role of micrornas in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 2015, 359, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. Microrna-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Pisco, A.; Papalopulu, N. Microrna-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 2011, 20, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.; Thieffry, D.; Drivenes, O.; Becker, T.S.; Bally-Cuif, L. Mir-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell 2012, 22, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Boissart, C.; Nissan, X.; Giraud-Triboult, K.; Peschanski, M.; Benchoua, A. Mir-125 potentiates early neural specification of human embryonic stem cells. Development 2012, 139, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.; Mendelson, J.; Blake, T.; Mishra, L.; Mishra, B. Tgf-beta signaling in neuronal stem cells. Dis. Mark. 2008, 24, 251–255. [Google Scholar] [CrossRef]
- Meyers, E.A.; Kessler, J.A. Tgf-beta family signaling in neural and neuronal differentiation, development, and function. Cold Spring Harb. Perspect. Biol. 2017, 9, a022244. [Google Scholar] [CrossRef] [PubMed]
- Inestrosa, N.C.; Varela-Nallar, L. Wnt signalling in neuronal differentiation and development. Cell Tissue Res. 2015, 359, 215–223. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.; Song, Y.; Kim, W.; Ying, Q.L.; Jho, E.H. Dual function of wnt signaling during neuronal differentiation of mouse embryonic stem cells. Stem Cells Int. 2015, 2015, 459301. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Matsuoka, A.J.; Shimomura, A.; Koehler, K.R.; Chan, R.J.; Miller, J.M.; Srour, E.F.; Hashino, E. Wnt signaling promotes neuronal differentiation from mesenchymal stem cells through activation of tlx3. Stem Cells 2011, 29, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Patton, B.; Eckley, D.M.; Magnus, T.; Mughal, M.R.; Sasaki, T.; Caldwell, M.A.; Rao, M.S.; Mattson, M.P.; ffrench-Constant, C. Patterns of laminins and integrins in the embryonic ventricular zone of the cns. J. Comp. Neurol. 2007, 505, 630–643. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Tavakoli, T.; Derby, E.; Serebryakova, Y.; Rao, M.S.; Mattson, M.P. Cell-extracellular matrix interactions regulate neural differentiation of human embryonic stem cells. BMC Dev. Biol. 2008, 8, 90. [Google Scholar] [CrossRef] [PubMed]






| Antibody | Dilution | Company Cat# and RRID |
|---|---|---|
| Rabbit anti-SOX1 | 1:200 | Cell Signaling Cat#9606 |
| Rabbit anti-SOX2 | 1:400 | Cell Signaling Cat#3579 |
| Mouse anti-Nestin | 1:500 | Stemcell Technologies Cat#60091 |
| Donkey Anti-Rabbit Alexa Fluor® 488 | 1:1000 | Jackson ImmunoResearch Cat#711-546-152 |
| Donkey Anti-Mouse Alexa Fluor® 594 | 1:1000 | Jackson ImmunoResearch Cat#715-586-151 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulcenty, K.; Wroblewska, J.P.; Rucinski, M.; Kozlowska, E.; Jopek, K.; Suchorska, W.M. MicroRNA Profiling During Neural Differentiation of Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2019, 20, 3651. https://doi.org/10.3390/ijms20153651
Kulcenty K, Wroblewska JP, Rucinski M, Kozlowska E, Jopek K, Suchorska WM. MicroRNA Profiling During Neural Differentiation of Induced Pluripotent Stem Cells. International Journal of Molecular Sciences. 2019; 20(15):3651. https://doi.org/10.3390/ijms20153651
Chicago/Turabian StyleKulcenty, Katarzyna, Joanna P Wroblewska, Marcin Rucinski, Emilia Kozlowska, Karol Jopek, and Wiktoria M Suchorska. 2019. "MicroRNA Profiling During Neural Differentiation of Induced Pluripotent Stem Cells" International Journal of Molecular Sciences 20, no. 15: 3651. https://doi.org/10.3390/ijms20153651