Rv0613c/MSMEG_1285 Interacts with HBHA and Mediates Its Proper Cell-Surface Exposure in Mycobacteria
Abstract
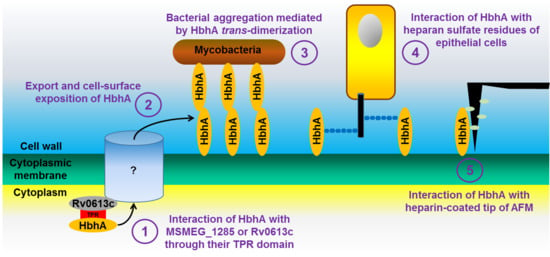
:1. Introduction
2. Results
2.1. The N-Terminal Part of HBHA Interacts with Rv0613c and MmpL14
2.2. Rv0613c Is Conserved within the Mtb Complex and Contains a SEC-C Motif
2.3. Generation of a ΔMSMEG_1285 Mutant in Msmeg mc2155
2.4. MSMEG_1285 Is Necessary for the Proper Cell-Surface Exposure of HBHA_Mtb
2.5. Loss of Auto-Aggregation in ΔMSMEG_1285 Expressing hbhA_Mtb
2.6. MSMEG_1285 Is Necessary for Full Infectivity of Msmeg mc2155 Expressing hbhA_Mtb
2.7. Proteomic Analysis of the ΔMSMEG_1285 Mutant
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Media
4.2. Generation of the ΔMSMEG_1285 Mutant in Msmeg mc2155
4.3. Plasmid Constructions
4.4. Bacterial Adenylate Cyclase Two-Hybrid Assays
4.5. Fractionation Protocol
4.6. Immunoblot Analysis
4.7. Atomic Force Microscopy
4.8. Infection of A549 Cells
4.9. Mass Spectrometry Proteomic Analysis
4.10. Proteomic Data Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- WHO. World Health Organization Global Tuberculosis Report; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Menozzi, F.D.; Rouse, J.H.; Alavi, M.; Laude-Sharp, M.; Muller, J.; Bischoff, R.; Brennan, M.J.; Locht, C. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 1996, 184, 993–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menozzi, F.D.; Bischoff, R.; Fort, E.; Brennan, M.J.; Locht, C. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc. Natl. Acad. Sci. USA 1998, 95, 12625–12630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.Y.; Irene, D.; Zulueta, M.M.; Tai, T.J.; Lain, S.H.; Cheng, C.P.; Tsai, P.X.; Lin, S.Y.; Chen, Z.G.; Ku, C.C.; et al. Structure of the Complex between a Heparan Sulfate Octasaccharide and Mycobacterial Heparin-Binding Hemagglutinin. Angew. Chem. Int. Ed. Engl. 2017, 56, 4192–4196. [Google Scholar] [CrossRef] [PubMed]
- Pethe, K.; Alonso, S.; Biet, F.; Delogu, G.; Brennan, M.J.; Locht, C.; Menozzi, F.D. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 2001, 412, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, N.; Saiga, H.; Houthuys, E.; Moura-Alves, P.; Koehler, A.; Bandermann, S.; Dorhoi, A.; Kaufmann, S.H. Syndecans promote mycobacterial internalization by lung epithelial cells. Cell. Microbiol. 2016, 18, 1846–1856. [Google Scholar] [CrossRef] [PubMed]
- Pethe, K.; Bifani, P.; Drobecq, H.; Sergheraert, C.; Debrie, A.S.; Locht, C.; Menozzi, F.D. Mycobacterial heparin-binding hemagglutinin and laminin-binding protein share antigenic methyllysines that confer resistance to proteolysis. Proc. Natl. Acad. Sci. USA 2002, 99, 10759–10764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temmerman, S.; Pethe, K.; Parra, M.; Alonso, S.; Rouanet, C.; Pickett, T.; Drowart, A.; Debrie, A.S.; Delogu, G.; Menozzi, F.D.; et al. Methylation-dependent T cell immunity to Mycobacterium tuberculosis heparin-binding hemagglutinin. Nat. Med. 2004, 10, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Biet, F.; Angela de Melo Marques, M.; Grayon, M.; Xavier da Silveira, E.K.; Brennan, P.J.; Drobecq, H.; Raze, D.; Vidal Pessolani, M.C.; Locht, C.; Menozzi, F.D. Mycobacterium smegmatis produces an HBHA homologue which is not involved in epithelial adherence. Microbes Infect. 2007, 9, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Lanfranconi, M.P.; Alvarez, H.M. Functional divergence of HBHA from Mycobacterium tuberculosis and its evolutionary relationship with TadA from Rhodococcus opacus. Biochimie 2016, 127, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.V.; Brosch, R.; Billault, A.; Garnier, T.; Eiglmeier, K.; Cole, S.T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 1999, 32, 643–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domenech, P.; Reed, M.B.; Barry, C.E., 3rd. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 2005, 73, 3492–3501. [Google Scholar] [CrossRef] [PubMed]
- Pethe, K.; Aumercier, M.; Fort, E.; Gatot, C.; Locht, C.; Menozzi, F.D. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. 2000, 275, 14273–14280. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, P.; Raze, D.; Fritzinger, B.; Wieruszeski, J.M.; Biet, F.; Dose, A.; Carpentier, M.; Schwarzer, D.; Allain, F.; Lippens, G.; et al. Differential contribution of the repeats to heparin binding of HBHA, a major adhesin of Mycobacterium tuberculosis. PLoS ONE 2012, 7, e32421. [Google Scholar] [CrossRef] [PubMed]
- Ortega, C.; Anderson, L.N.; Frando, A.; Sadler, N.C.; Brown, R.W.; Smith, R.D.; Wright, A.T.; Grundner, C. Systematic Survey of Serine Hydrolase Activity in Mycobacterium tuberculosis Defines Changes Associated with Persistence. Cell. Chem. Biol. 2016, 23, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Cerveny, L.; Straskova, A.; Dankova, V.; Hartlova, A.; Ceckova, M.; Staud, F.; Stulik, J. Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 2013, 81, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Dupres, V.; Menozzi, F.D.; Locht, C.; Clare, B.H.; Abbott, N.L.; Cuenot, S.; Bompard, C.; Raze, D.; Dufrene, Y.F. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2005, 2, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Raze, D.; Verwaerde, C.; Deloison, G.; Werkmeister, E.; Coupin, B.; Loyens, M.; Brodin, P.; Rouanet, C.; Locht, C. Heparin-binding Hemagglutinin Adhesin (HBHA) is involved in intracytosolic lipid inclusions (ILI) formation in mycobacteria. Manuscript in preparation. 2018. [Google Scholar]
- Delogu, G.; Brennan, M.J. Functional domains present in the mycobacterial hemagglutinin, HBHA. J. Bacteriol. 1999, 181, 7464–7469. [Google Scholar] [PubMed]
- Esposito, C.; Carullo, P.; Pedone, E.; Graziano, G.; Del Vecchio, P.; Berisio, R. Dimerisation and structural integrity of Heparin Binding Hemagglutinin A from Mycobacterium tuberculosis: Implications for bacterial agglutination. FEBS Lett. 2010, 584, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Mahamed, D.; Boulle, M.; Ganga, Y.; Mc Arthur, C.; Skroch, S.; Oom, L.; Catinas, O.; Pillay, K.; Naicker, M.; Rampersad, S.; et al. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. Elife 2017, 6, e22028. [Google Scholar] [PubMed]
- Noinaj, N.; Gumbart, J.C.; Buchanan, S.K. The beta-barrel assembly machinery in motion. Nat. Rev. Microbiol. 2017, 15, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, R.; Zeth, K. Structural basis of outer membrane protein biogenesis in bacteria. J. Biol. Chem. 2011, 286, 27792–27803. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, C.M.; Baker, S.L.; Jansen, K.; Metzner, S.I.; Sousa, M.C. Crystal structure of BamD: An essential component of the beta-Barrel assembly machinery of gram-negative bacteria. J. Mol. Biol. 2011, 409, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ohara, N.; Sato, K.; Yoshimura, M.; Yukitake, H.; Naito, M.; Fujiwara, T.; Nakayama, K. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 2010, 78, 2846–2856. [Google Scholar] [CrossRef] [PubMed]
- McGuire, A.M.; Weiner, B.; Park, S.T.; Wapinski, I.; Raman, S.; Dolganov, G.; Peterson, M.; Riley, R.; Zucker, J.; Abeel, T.; et al. Comparative analysis of Mycobacterium and related Actinomycetes yields insight into the evolution of Mycobacterium tuberculosis pathogenesis. BMC Genom. 2012, 13, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleischmann, R.D.; Alland, D.; Eisen, J.A.; Carpenter, L.; White, O.; Peterson, J.; DeBoy, R.; Dodson, R.; Gwinn, M.; Haft, D.; et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 2002, 184, 5479–5490. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Lu, L.; Wang, B.; Pu, S.; Zhang, X.; Zhu, G.; Shi, W.; Zhang, L.; Wang, H.; Wang, S.; et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS ONE 2008, 3, e2375. [Google Scholar] [CrossRef] [PubMed]
- Chalut, C. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis 2016, 100, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A.; Dubois, V.; Girard-Misguich, F.; Blaise, M.; Herrmann, J.L.; Kremer, L. The diverse family of MmpL transporters in mycobacteria: From regulation to antimicrobial developments. Mol. Microbiol. 2017, 104, 889–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chim, N.; Torres, R.; Liu, Y.; Capri, J.; Batot, G.; Whitelegge, J.P.; Goulding, C.W. The Structure and Interactions of Periplasmic Domains of Crucial MmpL Membrane Proteins from Mycobacterium tuberculosis. Chem. Biol. 2015, 22, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Bernut, A.; Viljoen, A.; Dupont, C.; Sapriel, G.; Blaise, M.; Bouchier, C.; Brosch, R.; de Chastellier, C.; Herrmann, J.L.; Kremer, L. Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol. Microbiol. 2016, 99, 866–883. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, J.C.; Marinelli, L.J.; Hatfull, G.F. Recombineering mycobacteria and their phages. Nat. Rev. Microbiol. 2008, 6, 851–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miguet, L.; Bechade, G.; Fornecker, L.; Zink, E.; Felden, C.; Gervais, C.; Herbrecht, R.; Van Dorsselaer, A.; Mauvieux, L.; Sanglier-Cianferani, S. Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J. Proteome Res. 2009, 8, 3346–3354. [Google Scholar] [CrossRef] [PubMed]





| Protein | MW | Fraction | mc2155 | ΔMSMEG_1285 | Complemented | mc2155/hbhA_Mtb | ΔMSMEG_1285/hbhA_Mtb |
|---|---|---|---|---|---|---|---|
| MSMEG_3496 | 106210 | MB | 1 (1) | 5 (4) | 1 (1) | 1 (1) | 14 (12) |
| CW | 4 (3) | 9 (6) | 4 (3) | 48 (22) | 15 (10) | ||
| MSMEG_1285 | 88003 | MB | 1 (1) | 0 | 4 (3) | 3 (3) | 0 |
| CW | 1 (1) | 0 | 11 (8) | 2 (2) | 0 | ||
| MSMEG_0919 (HBHA_Msmeg) | 24562 | MB | 28 (12) | 37 (13) | 27 (11) | 13 (7) | 23 (11) |
| CW | 15 (9) | 16 (10) | 16 (7) | 7 (5) | 13 (8) | ||
| Rv0475_EGFP (HBHA_Mtb) | 48472 | MB | 0 | 0 | 0 | 17 (14) | 14 (10) |
| CW | 0 | 0 | 0 | 4(4) | 5 (4) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veyron-Churlet, R.; Dupres, V.; Saliou, J.-M.; Lafont, F.; Raze, D.; Locht, C. Rv0613c/MSMEG_1285 Interacts with HBHA and Mediates Its Proper Cell-Surface Exposure in Mycobacteria. Int. J. Mol. Sci. 2018, 19, 1673. https://doi.org/10.3390/ijms19061673
Veyron-Churlet R, Dupres V, Saliou J-M, Lafont F, Raze D, Locht C. Rv0613c/MSMEG_1285 Interacts with HBHA and Mediates Its Proper Cell-Surface Exposure in Mycobacteria. International Journal of Molecular Sciences. 2018; 19(6):1673. https://doi.org/10.3390/ijms19061673
Chicago/Turabian StyleVeyron-Churlet, Romain, Vincent Dupres, Jean-Michel Saliou, Frank Lafont, Dominique Raze, and Camille Locht. 2018. "Rv0613c/MSMEG_1285 Interacts with HBHA and Mediates Its Proper Cell-Surface Exposure in Mycobacteria" International Journal of Molecular Sciences 19, no. 6: 1673. https://doi.org/10.3390/ijms19061673