Parkinson’s Disease: From Pathogenesis to Pharmacogenomics
Abstract
:1. Introduction
2. Pathogenic Mechanisms
3. Conventional Treatments
4. Pharmacogenomics
5. Novel Treatments
6. Further Considerations
Conflicts of Interest
References
- Von Campenhausen, S.; Bornschein, B.; Wick, R.; Bötzel, K.; Sampaio, C.; Poewe, W.; Oertel, W.; Siebert, U.; Berger, K.; Dodel, R. Prevalence and incidence of Parkinson’s disease in Europe. Eur. Neuropsychopharmacol. 2005, 15, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.M.; Liu, J.; Tian, Z.Y.; Lu, D.; Zhou, Y.Y. Systematic review of the prevalence and incidence of Parkinson’s disease in the People’s Republic of China. Neuropsychiatr. Dis. Treat. 2015, 15, 1467–1472. [Google Scholar] [CrossRef] [PubMed]
- Muangpaisan, W.; Hori, H.; Brayne, C. Systematic review of the prevalence and incidence of Parkinson’s disease in Asia. J. Epidemiol. 2009, 19, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, L.; Jette, N.; Frolkis, A.; Steeves, T.; Pringsheim, T. The Incidence of Parkinson’s Disease: A Systematic Review and Meta-Analysis. Neuroepidemiology 2016, 46, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Savica, R.; Grossardt, B.R.; Bower, J.H.; Ahlskog, J.E.; Rocca, W.A. Time Trends in the Incidence of Parkinson Disease. JAMA Neurol. 2016, 73, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Pringsheim, T.; Jette, N.; Frolkis, A.; Steeves, T.D. The prevalence of Parkinson’s disease: A systematic review and meta-analysis. Mov. Disord. 2014, 29, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Riedel, O.; Bitters, D.; Amann, U.; Garbe, E.; Langner, I. Estimating the prevalence of Parkinson’s disease (PD) and proportions of patients with associated dementia and depression among the older adults based on secondary claims data. Int. J. Geriatr. Psychiatry 2016, 31, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Moisan, F.; Kab, S.; Mohamed, F.; Canonico, M.; le Guern, M.; Quintin, C.; Carcaillon, L.; Nicolau, J.; Duport, N.; Singh-Manoux, A.; et al. Parkinson disease male-to-female ratios increase with age: French nationwide study and meta-analysis. J. Neurol. Neurosurg. Psychiatry 2016, 87, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R. World Guide for Drug Use and Pharmacogenomics; EuroEspes Publishing: Corunna, Spain, 2012. [Google Scholar]
- Cacabelos, R.; Cacabelos, P.; Torrellas, C.; Tellado, I.; Carril, J.C. Pharmacogenomics of Alzheimer’s disease: Novel therapeutic strategies for drug development. Methods Mol. Biol. 2014, 1175, 323–556. [Google Scholar] [PubMed]
- Cacabelos, R.; Torrellas, C.; Carrera, I. Opportunities in Pharmacogenomics for the treatment of Alzheimer’s Disease. Future Neurol. 2015, 10, 229–252. [Google Scholar] [CrossRef]
- Cacabelos, R.; Torrellas, C. Epigenetic drug discovery for Alzheimer's disease. Expert Opin. Drug Discov. 2014, 9, 1059–1086. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Torrellas, C. Epigenetics of aging and Alzheimer’s disease: Implications for pharmacogenomics and drug response. Int. J. Mol. Sci. 2015, 16, 30483–30543. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Torrellas, C.; Teijido, O.; Carril, J.C. Pharmacogenetic considerations in the treatment of Alzheimer’s disease. Pharmacogenomics 2016, 17, 1041–1074. [Google Scholar] [CrossRef] [PubMed]
- Katzenschlager, R.; Lees, A.J. Treatment of Parkinson’s disease: Levodopa as the first choice. J. Neurol. 2002, 249, II19–II24. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R. Parkinson’s disease: Old concepts and new challenges. Sci. Pages Alzheimers Dis. Dement. 2016, 1, 001. [Google Scholar]
- Cacabelos, R.; Fernández-Novoa, L.; Alejo, R.; Corzo, L.; Alcaraz, M.; Nebril, L.; Cacabelos, P.; Fraile, C.; Carrera, I.; Carril, J.C. E-PodoFavalin-15999 (Atremorine®)-induced dopamine response in Parkinson’s Disease: Pharmacogenetics-related effects. J. Genom. Med. Pharmacogenom. 2016, 1, 1–26. [Google Scholar]
- Cacabelos, R.; Fernández-Novoa, L.; Alejo, R.; Corzo, L.; Rodríguez, S.; Alcaraz, M.; Nebril, L.; Cacabelos, P.; Fraile, C.; Carrera, I.; et al. E-PodoFavalin-15999 (Atremorine®)-induced neurotransmitter and hormonal response in Parkinson’s Disease. J. Exp. Res. Pharmacol. 2016, 1, 1–12. [Google Scholar]
- Cacabelos, R.; Carrera, I.; Fernández-Novoa, L.; Alejo, R.; Corzo, L.; Rodríguez, S.; Alcaraz, M.; Nebril, L.; Casas, A.; Fraile, C.; et al. Parkinson’s Disease: New solutions to old problems. EuroEspes J. 2017, 11, 74–96. [Google Scholar]
- Miller, D.B.; O’Callaghan, J.P. Biomarkers of Parkinson’s disease: Present and future. Metabolism 2015, 64, S40–S46. [Google Scholar] [CrossRef] [PubMed]
- Naughton, C.; O’Toole, D.; Kirik, D.; Dowd, E. Interaction between subclinical doses of the Parkinson’s disease associated gene, α-synuclein, and the pesticide, rotenone, precipitates motor dysfunction and nigrostriatal neurodegeneration in rats. Behav. Brain Res. 2017, 316, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Ritz, B.R.; Paul, K.C.; Bronstein, J.M. Of Pesticides and Men: A California Story of Genes and Environment in Parkinson’s Disease. Curr. Environ. Health Rep. 2016, 3, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Rokad, D.; Ghaisas, S.; Harischandra, D.S.; Jin, H.; Anantharam, V.; Kanthasamy, A.; Kanthasamy, A.G. Role of Neurotoxicants and Traumatic Brain Injury in α-Synuclein Protein Misfolding and Aggregation. Brain Res. Bull. 2016. [Google Scholar] [CrossRef] [PubMed]
- Toledo, J.B.; Arnold, S.E.; Raible, K.; Brettschneider, J.; Xie, S.X.; Grossman, M.; Monsell, S.E.; Kukull, W.A.; Trojanowski, J.Q. Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer’s Coordinating Centre. Brain 2013, 136, 2697–2706. [Google Scholar] [CrossRef]
- Irwin, D.J.; Grossman, M.; Weintraub, D.; Hurtig, H.I.; Duda, J.E.; Xie, S.X.; Lee, E.B.; van Deerlin, V.M.; Lopez, O.L.; Kofler, J.K.; et al. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: A retrospective analysis. Lancet Neurol. 2017, 16, 55–65. [Google Scholar] [CrossRef]
- Wen, K.X.; Miliç, J.; El-Khodor, B.; Dhana, K.; Nano, J.; Pulido, T.; Kraja, B.; Zaciragic, A.; Bramer, W.M.; Troup, J.; et al. The Role of DNA Methylation and Histone Modifications in Neurodegenerative Diseases: A Systematic Review. PLoS ONE 2016, 11, e0167201. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, R.L. Genentics of synucleopathies. Cold Spring Harb. Perspect. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lill, C.M. Genetics of Parkinson’s disease. Mol. Cell. Probes 2016, 30, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Feng, H.; Peng, S.; Xiao, J.; Zhang, J. Association of plasma homocysteine, vitamin B12 and folate levels with cognitive function in Parkinson’s disease: A meta-analysis. Neurosci. Lett. 2017, 636, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Brundin, P. Parkinson’s disease and alpha synuclein: Is Parkinson’s disease a prion-like disorder? Mov. Disord. 2013, 28, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Scheffold, A.; Holtman, I.R.; Dieni, S.; Brouwer, N.; Katz, S.F.; Jebaraj, B.M.; Kahle, P.J.; Hengerer, B.; Lechel, A.; Stilgenbauer, S.; et al. Telomere shortening leads to an acceleration of synucleinopathy and impaired microglia response in a genetic mouse model. Acta Neuropathol. Commun. 2016, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Olszewska, D.A.; Fearon, C.; Lynch, T. Novel gene (TMEM230) linked to Parkinson’s disease. J. Clin. Mov. Disord. 2016, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Nuytemans, K.; Theuns, J.; Cruts, M.; van Broeckhoven, C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: A mutation update. Hum. Mutat. 2010, 31, 763–780. [Google Scholar] [CrossRef] [PubMed]
- Lardenoije, R.; Iatrou, A.; Kenis, G.; Kompotis, K.; Steinbusch, H.W.; Mastroeni, D.; Coleman, P.; Lemere, C.A.; Hof, P.R.; van den Hove, D.L.; et al. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015, 131, 21–64. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F. Genetics and epigenetics of Parkinson’s disease. Sci. World J. 2012. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.G.; Reed, X.; Singleton, A.B. Genetics in Parkinson’s disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016, 139, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Hill-Burns, E.M.; Ross, O.A.; Wissemann, W.T.; Soto-Ortolaza, A.I.; Zareparsi, S.; Siuda, J.; Lynch, T.; Wszolek, Z.K.; Silburn, P.A.; Mellick, G.D.; et al. Identification of genetic modifiers of age-at-onset for familial Parkinson’s disease. Hum. Mol. Genet. 2016, 25, 3849–3862. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, R. Phenome-based gene discovery provides information about Parkinson’s disease drug targets. BMC Genom. 2016, 17, 493. [Google Scholar] [CrossRef] [PubMed]
- Sandor, C.; Honti, F.; Haerty, W.; Szewczyk-Krolikowski, K.; Tomlinson, P.; Evetts, S.; Millin, S.; Keane, T.; McCarthy, S.A.; Durbin, R.; et al. Whole-exome sequencing of 228 patients with sporadic Parkinson’s disease. Sci. Rep. 2017, 7, 41188. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, A.; Theuns, J.; van Broeckhoven, C. Progress in unraveling the genetic etiology of Parkinson disease in a genomic era. Trends Genet. 2015, 31, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Lill, C.M.; Roehr, J.T.; McQueen, M.B.; Kavvoura, F.K.; Bagade, S.; Schjeide, B.M.; Schjeide, L.M.; Meissner, E.; Zauft, U.; Allen, N.C.; et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012, 8, e1002548. [Google Scholar] [CrossRef] [PubMed]
- Do, C.B.; Tung, J.Y.; Dorfman, E.; Kiefer, A.K.; Drabant, E.M.; Francke, U.; Mountain, J.L.; Goldman, S.M.; Tanner, C.M.; Langston, J.W.; et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011, 7, e1002141. [Google Scholar] [CrossRef] [PubMed]
- International Parkinson Disease Genomics Consortium; Nalls, M.A.; Plagnol, V.; Hernandez, D.G.; Sharma, M.; Sheerin, U.M.; Saad, M.; Simón-Sánchez, J.; Schulte, C.; Lesage, S.; et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet 2011, 377, 641–649. [Google Scholar]
- Edwards, T.L.; Scott, W.K.; Almonte, C.; Burt, A.; Powell, E.H.; Beecham, G.W.; Wang, L.; Züchner, S.; Konidari, I.; Wang, G.; et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010, 74, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Pankratz, N.; Wilk, J.B.; Latourelle, J.C.; DeStefano, A.L.; Halter, C.; Pugh, E.W.; Doheny, K.F.; Gusella, J.F.; Nichols, W.C.; Foroud, T.; et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 2009, 124, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Pankratz, N.; Beecham, G.W.; de Stefano, A.L.; Dawson, T.M.; Doheny, K.F.; Factor, S.A.; Hamza, T.H.; Hung, A.Y.; Hyman, B.T.; Ivinson, A.J.; et al. Meta-analysis of Parkinson’s disease: Identification of a novel locus, RIT2. Ann. Neurol. 2012, 71, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Simón-Sánchez, J.; van Hilten, J.J.; van de Warrenburg, B.; Post, B.; Berendse, H.W.; Arepalli, S.; Hernandez, D.G.; de Bie, R.M.; Velseboer, D.; Scheffer, H.; et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur. J. Hum. Genet. 2011, 19, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, R.; Verbitsky, M.; Kisselev, S.; Browne, A.; Mejia-Sanatana, H.; Louis, E.D.; Cote, L.J.; Andrews, H.; Waters, C.; et al. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med. Genet. 2011, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.G.; Nalls, M.A.; Ylikotila, P.; Keller, M.; Hardy, J.A.; Majamaa, K.; Singleton, A.B. Genome wide assessment of young onset Parkinson’s disease from Finland. PLoS ONE 2012, 7, e41859. [Google Scholar] [CrossRef] [PubMed]
- Foo, J.N.; Tan, L.C.; Irwan, I.D.; Au, W.L.; Low, H.Q.; Prakash, K.M.; Ahmad-Annuar, A.; Bei, J.; Chan, A.Y.; Chen, C.M.; et al. Genome-wide association study of Parkinson’s disease in East Asians. Hum. Mol. Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Mu, H.; Shang, Z.; Kang, K.; Lv, H.; Duan, L.; Li, J.; Chen, X.; Teng, Y.; Jiang, Y.; et al. Genome-wide pathway-based association analysis identifies risk pathways associated with Parkinson’s disease. Neuroscience 2017, 340, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cen, L.; Xiao, Y.; Wei, L.; Mo, M.; Chen, X.; Li, S.; Yang, X.; Huang, Q.; Qu, S.; Pei, Z.; et al. Association of DYRK1A polymorphisms with sporadic Parkinson’s disease in Chinese Han population. Neurosci. Lett. 2016, 632, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Funayama, M.; Ohe, K.; Amo, T.; Furuya, N.; Yamaguchi, J.; Saiki, S.; Li, Y.; Ogaki, K.; Ando, M.; Yoshino, H.; et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: A genome-wide linkage and sequencing study. Lancet Neurol. 2015, 14, 274–282. [Google Scholar] [CrossRef]
- Mohan, M.; Mellick, G.D. Role of the VPS35 D620N mutation in Parkinson’s disease. Park. Relat. Disord. 2016. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.H.; Zhang, S.Y.; Yang, Z.H.; Yang, J.; Shang, D.D.; Mao, C.Y.; Liu, H.; Hou, H.M.; Shi, MM.; Wu, J.; et al. A novel RAB39B gene mutation in X-linked juvenile Parkinsonism with basal ganglia calcification. Mov. Disord. 2016, 31, 1905–1909. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.X.; Shi, Y.; Yang, Y.; Ahmeti, K.B.; Miller, N.; Huang, C.; Cheng, L.; Zhai, H.; Deng, S.; Nuytemans, K.; et al. Identification of TMEM230 mutations in familial Parkinson’s disease. Nat. Genet. 2016, 48, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, L.; Li, N.N.; Yu, W.J.; Sun, X.Y.; Li, J.Y.; Zhou, D.; Peng, R. SNCA rs356182 variant increases risk of sporadic Parkinson’s disease in ethnic Chinese. J. Neurol. Sci. 2016, 368, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Migdalska-Richards, A.; Schapira, A.H. The relationship between glucocerebrosidase mutations and Parkinson disease. J. Neurochem. 2016, 139, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Ferrazza, R.; Cogo, S.; Melrose, H.; Bubacco, L.; Greggio, E.; Guella, G.; Civiero, L.; Plotegher, N. LRRK2 deficiency impacts ceramide metabolism in brain. Biochem. Biophys. Res. Commun. 2016, 478, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mo, M.; Li, G.; Cen, L.; Wei, L.; Xiao, Y.; Chen, X.; Li, S.; Yang, X.; Qu, S.; et al. The biomarkers of immune dysregulation and inflammation response in Parkinson disease. Transl. Neurodegener. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yu, S.; Zheng, Y.; Yang, H.; Zhang, J. Oxidative Modification and Its Implications for the Neurodegeneration of Parkinson’s Disease. Mol. Neurobiol. 2017, 54, 1404–1418. [Google Scholar] [CrossRef] [PubMed]
- Kleinknecht, A.; Popova, B.; Lázaro, D.F.; Pinho, R.; Valerius, O.; Outeiro, T.F.; Braus, G.H. C-Terminal Tyrosine Residue Modifications Modulate the Protective Phosphorylation of Serine 129 of α-Synuclein in a Yeast Model of Parkinson’s Disease. PLoS Genet. 2016, 12, e1006098. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.; Kim, K.S.; Seol, W.; Choi, H.J. LRRK2 interferes with aggresome formation for autophagic clearance. Mol. Cell. Neurosci. 2016, 75, 71–80. [Google Scholar] [CrossRef]
- Zhang, C.W.; Hang, L.; Yao, T.P.; Lim, K.L. Parkin Regulation and Neurodegenerative Disorders. Front. Aging Neurosci. 2016, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Beal, M.F. Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 2016, 1802, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Buhlman, L.M. Parkin loss-of-function pathology: Premature neuronal senescence induced by high levels of reactive oxygen species? Mech. Ageing Dev. 2016, 161, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.D.; Sathiyamoorthy, S.; Angeles, D.C.; Tan, E.K. Linking F-box protein 7 and parkin to neuronal degeneration in Parkinson’s disease (PD). Mol. Brain 2016, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.R.; Randle, S.J.; Patel, S.P.; Mevissen, T.E.; Zenkeviciute, G.; Koide, T.; Komander, D.; Laman, H. Gsk3β and Tomm20 are substrates of the SCFFbxo7/PARK15 ubiquitin ligase associated with Parkinson’s disease. Biochem. J. 2016, 473, 3563–3580. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Akazawa-Ogawa, Y.; Matsumura, A.; Saigoh, K.; Itoh, S.; Sutou, K.; Kobayashi, M.; Mita, Y.; Shichiri, M.; Hisahara, S.; et al. Oxidation and interaction of DJ-1 with 20S proteasome in the erythrocytes of early stage Parkinson’s disease patients. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Mo, M.S.; Huang, W.; Sun, C.C.; Zhang, L.M.; Cen, L.; Xiao, Y.S.; Li, G.F.; Yang, X.L.; Qu, S.G.; Xu, P.Y. Association Analysis of Proteasome Subunits and Transporter Associated with Antigen Processing on Chinese Patients with Parkinson’s Disease. Chin. Med. J. 2016, 129, 1053–1058. [Google Scholar] [PubMed]
- Pihlstrøm, L.; Berge, V.; Rengmark, A.; Toft, M. Parkinson’s disease correlates with promoter methylation in the α-synuclein gene. Mov. Disord. 2015, 30, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Ammal Kaidery, N.; Tarannum, S.; Thomas, B. Epigenetic landscape of Parkinson’s disease: Emerging role in disease mechanisms and therapeutic modalities. Neurotherapeutics 2013, 10, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Masliah, E.; Dumaop, W.; Galasko, D.; Desplats, P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; McKnight, A.J.; Craig, D.; O’Neill, F. Epigenome-wide association study for Parkinson’s disease. Neuromol. Med. 2014, 16, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Torrellas, C.; Carril, J.C.; Aliev, G.; Teijido, O. Epigenomics and proteomics of brain disorders. Curr. Genom. 2017, in press. [Google Scholar]
- Mo, M.; Xiao, Y.; Huang, S.; Cen, L.; Chen, X.; Zhang, L.; Luo, Q.; Li, S.; Yang, X.; Lin, X.; et al. MicroRNA expressing profiles in A53T mutant alpha-synuclein transgenic mice and Parkinsonian. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Oertel, W.; Schulz, J.B. Current and experimental treatments of Parkinson disease: A guide for neuroscientists. J. Neurochem. 2016, 139, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Pahwa, R.; Lyons, K.E. Levodopa-related wearing-off in Parkinson’s disease: Identification and management. Curr. Med. Res. Opin. 2009, 25, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Bhidayasiri, R.; Hattori, N.; Jeon, B.; Chen, R.S.; Lee, M.K.; Bajwa, J.A.; Mok, V.C.; Zhang, B.; Syamsudin, T.; Tan, L.C.; et al. Asian perspectives on the recognition and management of levodopa ‘wearing-off’ in Parkinson’s disease. Expert Rev. Neurother. 2015, 15, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Haaxma, C.A.; Horstink, M.W.; Zijlmans, J.C.; Lemmens, W.A.; Bloem, B.R.; Borm, G.F. Risk of disabling response fluctuations and dyskinesias for dopamine agonists versus Levodopa in Parkinson’s disease. J. Parkinsons Dis. 2015, 5, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Rascol, O.; Perez-Lloret, S.; Ferreira, J.J. New treatments for levodopa-induced motor complications. Mov. Disord. 2015, 30, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Stowe, R.; Ives, N.; Clarke, C.E.; Deane, K.; van Hilten, J.; Wheatley, K.; Gray, R.; Handley, K.; Furmston, A. Evaluation of the efficacy and safety of adjuvant treatment to levodopa therapy in Parkinson s disease patients with motor complications. Cochrane Database Syst. Rev. 2010, 7, CD007166. [Google Scholar]
- Lertxundi, U.; Isla, A.; Solinis, M.A.; Domingo-Echaburu, S.; Hernandez, R.; Peral-Aguirregoitia, J.; Medrano, J. Anticholinergic burden in Parkinson’s disease inpatients. Eur. J. Clin. Pharmacol. 2015, 71, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Owolabi, L.F.; Samaila, A.A.; Sunmonu, T. Gastrointestinal complications in newly diagnosed Parkinson’s disease: A case-control study. Trop. Gastroenterol. 2014, 35, 227–231. [Google Scholar] [PubMed]
- Knudsen, K.; Krogh, K.; Østergaard, K.; Borghammer, P. Constipation in Parkinson’s disease: Subjective symptoms, objective markers, and new perspectives. Mov. Disord. 2017, 32, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Brophy, J.M.; Suissa, S.; Renoux, C. Risks of Cardiac Valve Regurgitation and Heart Failure Associated with Ergot- and Non-Ergot-Derived Dopamine Agonist Use in Patients with Parkinson’s Disease: A Systematic Review of Observational Studies. CNS Drugs 2015, 29, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Nakamura, T.; Hirayama, M.; Ueda, M.; Katsuno, M.; Sobue, G. Cardiac parasympathetic dysfunction in the early phase of Parkinson’s disease. J. Neurol. 2016, 264, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Barone, P.; Santangelo, G.; Amboni, M.; Pellecchia, M.T.; Vitale, C. Pisa syndrome in Parkinson’s disease and Parkinsonism: Clinical features, pathophysiology, and treatment. Lancet Neurol. 2016, 15, 1063–1074. [Google Scholar] [CrossRef]
- Altmann, V.; Schumacher-Schuh, A.F.; Rieck, M.; Callegari-Jacques, S.M.; Rieder, C.R.; Hutz, M.H. Influence of genetic, biological and pharmacological factors on levodopa dose in Parkinson’s disease. Pharmacogenomics 2016, 17, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Jiménez, F.J.; Alonso-Navarro, H.; García-Martín, E.; Agúndez, J.A. Advances in understanding genomic markers and pharmacogenetics of Parkinson’s disease. Expert Opin. Drug Metab. Toxicol. 2016, 12, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Kurzawski, M.; Białecka, M.; Droździk, M. Pharmacogenetic considerations in the treatment of Parkinson’s disease. Neurodegener. Dis. Manag. 2015, 5, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Schumacher-Schuh, A.F.; Rieder, C.R.; Hutz, M.H. Parkinson’s disease pharmacogenomics: New findings and perspectives. Pharmacogenomics 2014, 15, 1253–1271. [Google Scholar] [CrossRef] [PubMed]
- Rieck, M.; Schumacher-Schuh, A.F.; Callegari-Jacques, S.M.; Altmann, V.; Schneider Medeiros, M.; Hutz, M.H. Is there a role for ADORA2A polymorphisms in levodopa-induced dyskinesia in Parkinson’s disease patients? Pharmacogenomics 2015, 16, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Schumacher-Schuh, A.F.; Altmann, V.; Rieck, M.; Tovo-Rodrigues, L.; Monte, T.L.; Callegari-Jacques, S.M.; Medeiros, M.S.; Rieder, C.R.; Hutz, M.H. Association of common genetic variants of HOMER1 gene with levodopa adverse effects in Parkinson’s disease patients. Pharmacogenom. J. 2014, 14, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Rieck, M.; Schumacher-Schuh, A.F.; Altmann, V.; Francisconi, C.L.; Fagundes, P.T.; Monte, T.L.; Callegari-Jacques, S.M.; Rieder, C.R.; Hutz, M.H. DRD2 haplotype is associated with dyskinesia induced by levodopa therapy in Parkinson’s disease patients. Pharmacogenomics 2012, 13, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Moreau, C.; Meguig, S.; Corvol, J.C.; Labreuche, J.; Vasseur, F.; Duhamel, A.; Delval, A.; Bardyn, T.; Devedjian, J.C.; Rouaix, N.; et al. Polymorphism of the dopamine transporter type 1 gene modifies the treatment response in Parkinson’s disease. Brain 2015, 138, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.S.; Husain, R.S.; Kumar, S.; Ramakrishnan, V. Association between MDR1 gene polymorphisms and Parkinson’s disease in Asian and Caucasian populations: A meta-analysis. J. Neurol. Sci. 2016, 368, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Lohr, K.-M.; Masoud, S.T.; Salahpour, A.; Miller, G.W. Membrane transporters as mediators of synaptic dopamine dynamics: Implications for disease. Eur. J. Neurosci. 2017, 45, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jeon, B.; Chung, S.J. Professional ethics in complementary and alternative medicines in management of Parkinson’s disease. J. Park. Dis. 2016, 6, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Carrera, I.; Fernández-Novoa, L.; Sampedro, C.; Cacabelos, R. Neuroprotective effect of atremorine in an experimental model of Parkinson’s disease. Curr. Pharm. Des. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R. Bioactive Extract Obtained from Vicia Faba and Its Use in the Treatment and/or Prevention of Neurodegenerative Diseases. European Patent EP16382138, 29 March 2016. [Google Scholar]
- Goodarzi, Z.; Mele, B.; Guo, A.; Hanson, H.; Jette, N.; Patten, S.; Pringsheim, T.; Holroyd-Leduc, J. Guidelines for dementia or Parkinson’s disease with depression or anxiety: A systemic review. BMC Neurol. 2016, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, J.R.; Pontes, A.L.; Fiuza, F.P.; Silva, K.D.; Guzen, F.P.; Lucena, E.E.; Nascimento-Júnior, E.S.; Cavalcante, J.C.; Costa, M.S.; Engelberth, R.C.; et al. Nuclear organization of the substantia nigra, ventral tegmental area and retrorubral field of the common marmoset (Callithrix jacchus): A cytoarchitectonic and TH-immunohistochemistry study. J. Chem. Neuroanat. 2016, 77, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Sulzer, D.; Cragg, S.J.; Rice, M.E. Striatal dopamine neurotransmission: Regulation of release and uptake. Basal Ganglia 2016, 6, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Chandler, D.J.; Waterhouse, B.D.; Gao, W.J. New perspectives on catecholaminergic regulation of executive circuits: Evidence for independent modulation of prefrontal functions by midbrain dopaminergic and noradrenergic neurons. Front. Neural Circuits 2014, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Xing, B.; Li, Y.C.; Gao, W.J. Norepinephrine versus dopamine and their interaction in modulating synaptic function in the prefrontal cortex. Brain Res. 2016, 1641, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, K.; Tabuchi, K.; Ojika, M.; Zucca, F.A.; Zecca, L.; Ito, S. Norepinephrine and its metabolites are involved in the synthesis of neuromelanin derived from the locus coeruleus. J. Neurochem. 2015, 135, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Buddhala, C.; Loftin, S.K.; Kuley, B.M.; Cairns, N.J.; Campbell, M.C.; Perlmutter, J.S.; Joel, S. Dopaminergic, serotonergic, and noradrenergic deficits in Parkinson disease. Ann. Clin. Transl. Neurol. 2015, 2, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. Post mortem studies in Parkinson’s disease—Is it possible to detect brain areas for specific symptoms? J. Neural Transm. Suppl. 1999, 56, 1–29. [Google Scholar] [PubMed]
- Grouleff, J.; Ladefoged, L.K.; Koldsø, H.; Schiøtt, B. Monoamine transporters: Insights from molecular dynamics simulations. Front. Pharmacol. 2015, 6, 235. [Google Scholar] [CrossRef] [PubMed]
- Lohr, K.M.; Bernstein, A.I.; Stout, K.A.; Dunn, A.R.; Lazo, C.R.; Alter, S.P.; Wang, M.; Li, Y.; Fan, X.; Hess, E.J.; et al. Increased vesicular monoamine transporter enhances dopamine release and opposes Parkinson disease-related neurodegeneration in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 9977–9982. [Google Scholar] [CrossRef] [PubMed]
- Nagatsu, T.; Nagatsu, I. Tyrosine hydroxylase (TH), its cofactor tetrahydrobiopterin (BH4), other catecholamine-related enzymes, and their human genes in relation to the drug and gene therapies of Parkinson’s disease (PD): Historical overview and future prospects. J. Neural Transm. 2016, 123, 1255–1278. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.M.; Meadows, S.M.; Melikhov-Sosin, M.; Lindenbach, D.; Hallmark, J.; Werner, D.F. Monoamine transporter contributions to l-DOPA effects in hemi-parkinsonian rats. Neuropharmacology 2016, 110, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Debs, L.H.; Patel, A.P.; Nguyen, D.; Patel, K.; O’Connor, G.; Grati, M.; Mittal, J.; Yan, D.; Eshraghi, A.A.; et al. Neurotransmitters: The Critical Modulators Regulating Gut-Brain Axis. J. Cell. Physiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Cenci, M.A. Presynaptic Mechanisms of l-DOPA-Induced Dyskinesia: The Findings, the Debate, and the Therapeutic Implications. Front. Neurol. 2014, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Goldstein, D.S. Cardiovascular dysautonomia in Parkinson disease: From pathophysiology to pathogenesis. Neurobiol. Dis. 2012, 46, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W. Levodopa: Effect on cell death and the natural history of Parkinson’s disease. Mov. Disord. 2015, 30, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Nagatsu, T.; Sawada, M. Biochemistry of postmortem brains in Parkinson’s disease: Historical overview and future prospects. J. Neural Transm. Suppl. 2007, 72, 113–120. [Google Scholar]
- Vermeiren, Y.; de Deyn, P.P. Targeting the norepinephrinergic system in Parkinson’s disease and related disorders: The locus coeruleus story. Neurochem. Int. 2016, 102, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, D.L.; Kalinin, S.; Braun, D. Causes, consequences, and cures for neuroinflammation mediated via the locus coeruleus: Noradrenergic signaling system. J. Neurochem. 2016, 139, 154–178. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.; Madrigal, J.L.; Feinstein, D.L. Noradrenergic regulation of glial activation: Molecular mechanisms and therapeutic implications. Curr. Neuropharmacol. 2014, 12, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Espay, A.J.; LeWitt, P.A.; Kaufmann, H. Norepinephrine deficiency in Parkinson’s disease: The case for noradrenergic enhancement. Mov. Disord. 2014, 29, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Rommelfanger, K.S.; Weinshenker, D. Norepinephrine: The redheaded stepchild of Parkinson’s disease. Biochem. Pharmacol. 2007, 74, 177–190. [Google Scholar] [CrossRef]
- Mejias-Aponte, C.A. Specificity and impact of adrenergic projections to the midbrain dopamine system. Brain Res. 2016, 1641, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Aziz, N.A.; Pijl, H.; Frölich, M.; Roelfsema, F.; Roos, R.A. Diurnal secretion profiles of growth hormone, thyrotrophin and prolactin in Parkinson’s disease. J. Neuroendocrinol. 2011, 23, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Willis, G.L. Parkinson’s disease as a neuroendocrine disorder of circadian function: Dopamine-melatonin imbalance and the visual system in the genesis and progression of the degenerative process. Rev. Neurosci. 2008, 19, 245–316. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, S.; Vogt, T.; Nowak, T.; Kann, P.H. German KIMS board. Pituitary function and the somatotrophic system in patients with idiopathic Parkinson’s disease under chronic dopaminergic therapy. J. Neuroendocrinol. 2008, 20, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, A.; Ren, S.G.; Dong, J.; Culler, M.D.; Melmed, S. Regulation of growth hormone and prolactin gene expression and secretion by chimeric somatostatin-dopamine molecules. Endocrinology 2007, 148, 6107–6114. [Google Scholar] [CrossRef] [PubMed]
- Wells, S.; Murphy, D. Transgenic studies on the regulation of the anterior pituitary gland function by the hypothalamus. Front. Neuroendocrinol. 2003, 24, 11–26. [Google Scholar] [CrossRef]
- Jin, J.; Hara, S.; Sawai, K.; Fülöp, F.; Nagy, G.M.; Hashizume, T. Effects of hypothalamic dopamine (DA) on salsolinol (SAL)-induced prolactin (PRL) secretion in male goats. Anim. Sci. J. 2014, 85, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Nitkowska, M.; Tomasiuk, R.; Czyżyk, M.; Friedman, A. Prolactin and sex hormones levels in males with Parkinson’s disease. Acta Neurol. Scand. 2015, 131, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Kimber, J.; Watson, L.; Mathias, C.J. Neuroendocrine responses to levodopa in multiple system atrophy (MSA). Mov. Disord. 1999, 14, 981–987. [Google Scholar] [CrossRef]
- Kimber, J.R.; Watson, L.; Mathias, C.J. Distinction of idiopathic Parkinson’s disease from multiple-system atrophy by stimulation of growth-hormone release with clonidine. Lancet 1997, 349, 1877–1881. [Google Scholar] [CrossRef]
- Winkler, A.S.; Landau, S.; Chaudhuri, K.R. Serum prolactin levels in Parkinson’s disease and multiple system atrophy. Clin. Auton. Res. 2002, 12, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Kanazawa, M.; Takayanagi, M.; Kitani, Y.; Shintaku, H.; Kohno, Y. A case of 6-pyruvoyl-tetrahydropterin synthase deficiency demonstrates a more significant correlation of l-Dopa dosage with serum prolactin levels than CSF homovanillic acid levels. Brain Dev. 2008, 30, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Welnic, J.; Woitalla, D.; Muhlack, S. Endurance exercise modulates levodopa induced growth hormone release in patients with Parkinson’s disease. Neurosci. Lett. 2007, 422, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Kasuya, E.; Yayou, K.; Sutoh, M. L-DOPA attenuates prolactin secretion in response to isolation stress in Holstein steers. Anim. Sci. J. 2013, 84, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Székács, D.; Bodnár, I.; Mravec, B.; Kvetnansky, R.; Vizi, E.S.; Nagy, G.M.; Fekete, M.I. The peripheral noradrenergic terminal as possible site of action of salsolinol as prolactoliberin. Neurochem. Int. 2007, 50, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Levandis, G.; Balestra, B.; Siani, F.; Rizzo, V.; Ghezzi, C.; Ambrosi, G. Response of colonic motility to dopaminergic stimulation is subverted in rats with nigrostriatal lesion: Relevance to gastrointestinal dysfunctions in Parkinson’s disease. Neurogastroenterol. Motil. 2015, 27, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R. Biochemical changes and cardiovascular function in Parkinson’s disease. Clin. Med. Biochem. 2016, 2, 2. [Google Scholar] [CrossRef]
- Zhang, Z.; Du, X.; Xu, H.; Xie, J.; Jiang, H. Lesion of medullary catecholaminergic neurons is associated with cardiovascular dysfunction in rotenone-induced Parkinson’s disease rats. Eur. J. Neurosci. 2015, 42, 2346–2355. [Google Scholar] [CrossRef] [PubMed]
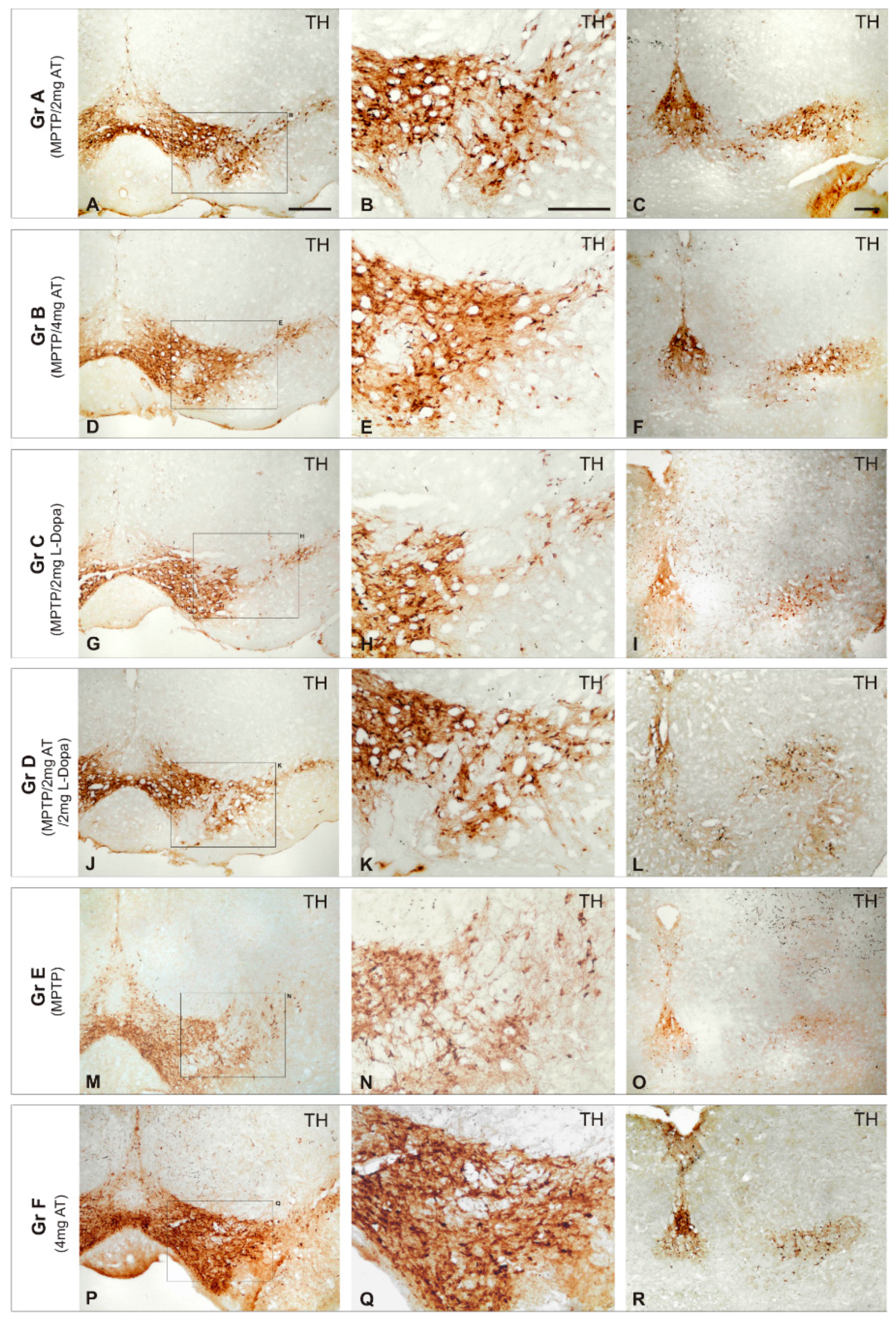

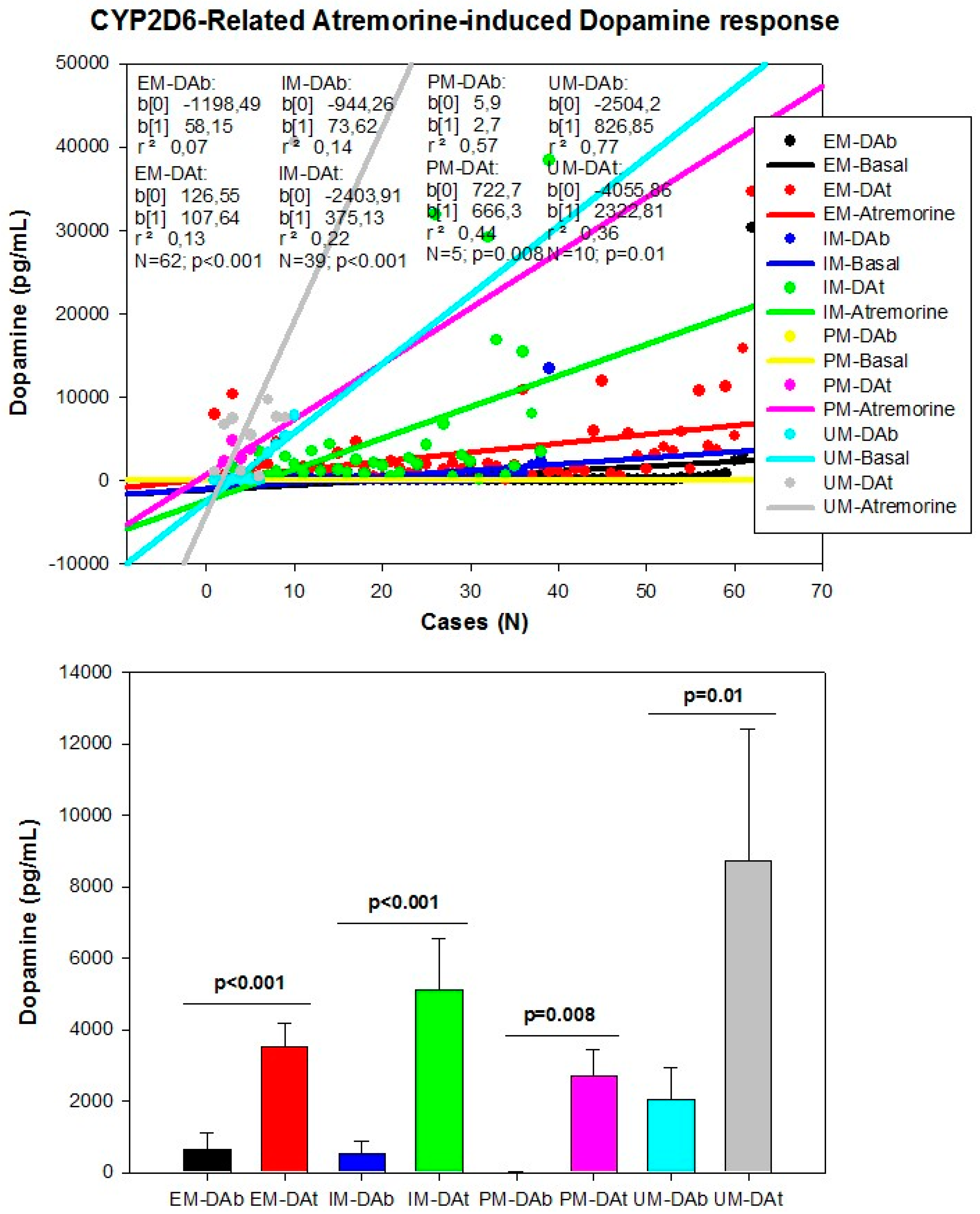
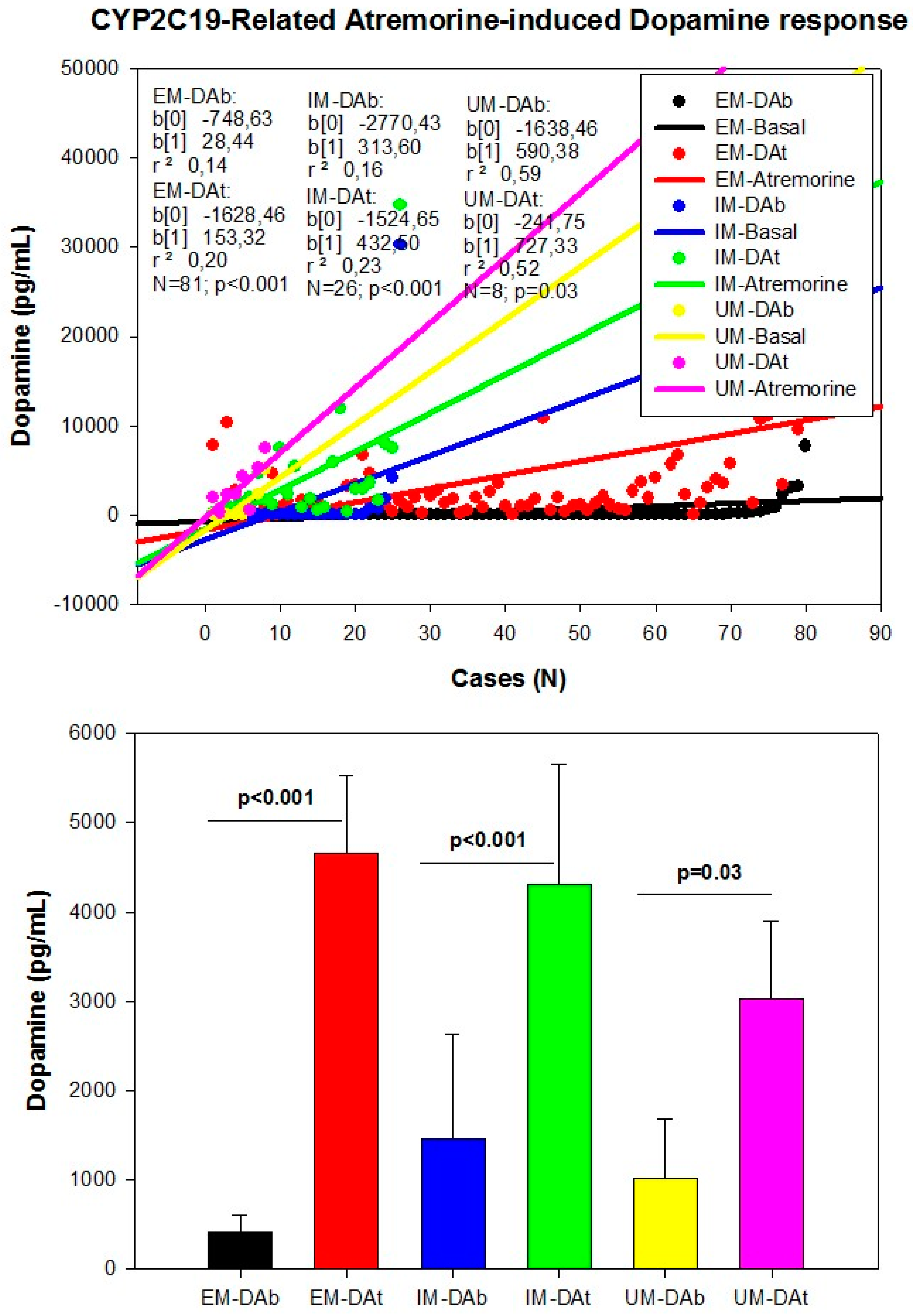
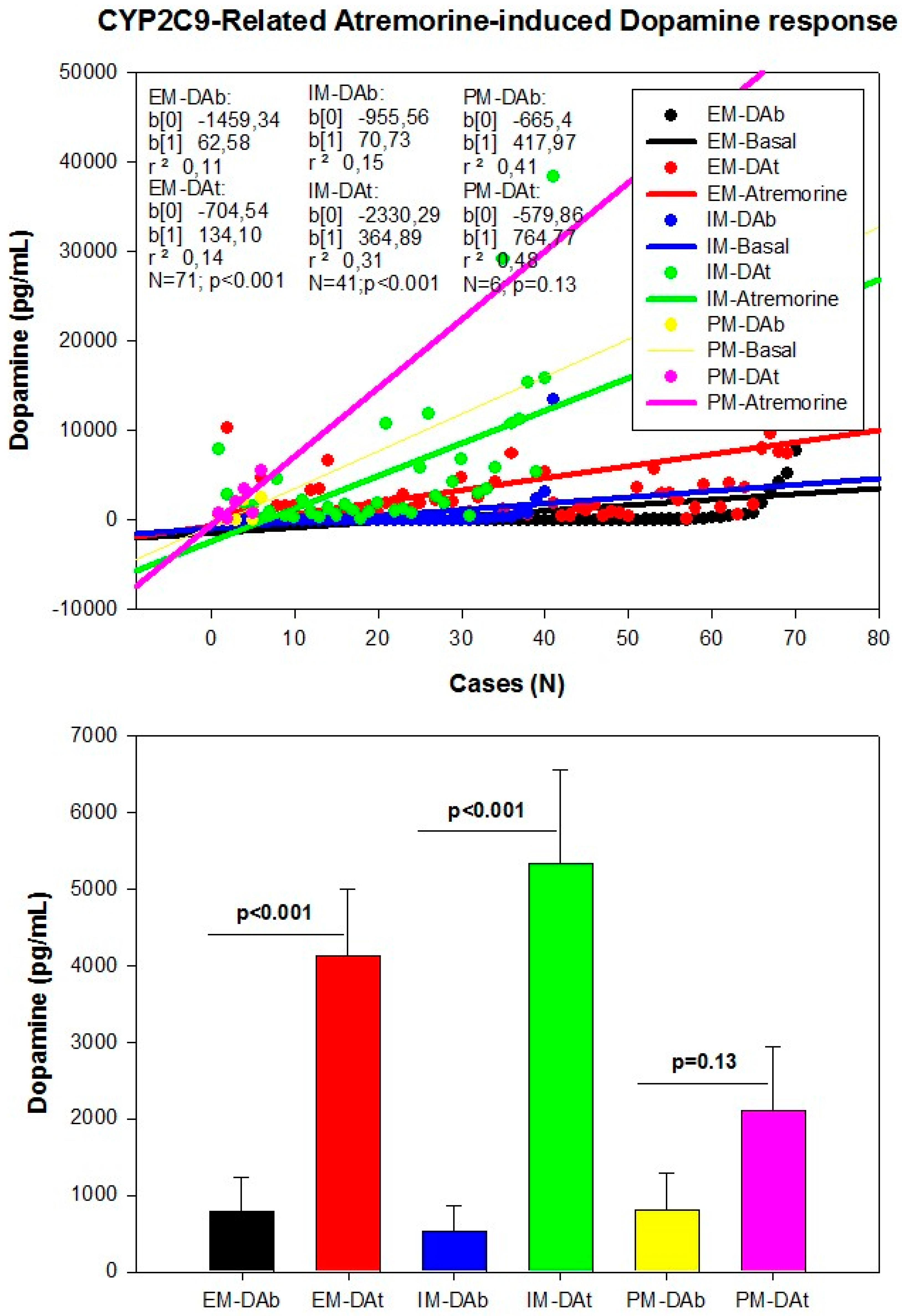
| Dopamine Precursors | ||
| Drug | Properties | Pharmacogenetics |
 | Name: Carbidopa; 28860-95-9; Lodosyn. IUPAC Name: Benzenepropanoic acid, α-hydrazino-3,4-dihydroxy-α-methyl-, monohydrate,(S) Molecular Formula: C10H14N2O4 H2O Molecular Weight: 244.24 g/mol Mechanism: Carbidopa is a peripheral decarboxylase inhibitor with little or no pharmacological activity when given alone in usual doses. It inhibits the peripheral decarboxylation of levodopa to dopamine. At the same time, reduced peripheral formation of dopamine reduces peripheral side effects, notably nausea or vomiting, and cardiac arrhythmias, although the dyskinesias and adverse mental effects associated with levodopa therapy tend to develop earlier. Effect: Antiparkinsonian agents. Dopamine precursors. | Pathogenic genes: BDNF, PARK2 Mechanistic genes: DRD2, OPRM1 Metabolic genes: Substrate: COMT, DDC Pleiotropic genes: ACE, ACHE |
 | Name: Levodopa; 59-92-7; Levodopa; l-DOPA; Dopar; Bendopa; Dopasol; 3,4-dihydroxy-l-phenylalanine; Madopar. IUPAC Name: l-Tyrosine-3-hydroxy Molecular Formula: C9H11NO4 Molecular Weight: 197.19 g/mol Mechanism: Levodopa circulates in the plasma to the blood–brain–barrier, where it crosses, to be converted by striatal enzymes to dopamine. Carbidopa inhibits the peripheral plasma breakdown of levodopa by inhibiting its carboxylation, and thereby increases available levodopa at the blood–brain–barrier. Effect: Antiparkinsonian agents. Dopamine precursors. | Pathogenic genes: ANKK1, BDNF, LRRK2, PARK2 Mechanistic genes: CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2, CYP2B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DBH, DDC, G6PD, MAOB, TH, UGT1A1, UGT1A9 Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, ACHE |
| Dopaminergic Agonists | ||
| Drug | Properties | Pharmacogenetics |
 | Name: Amantadine; 768-94-5; Amantadine; Symmetrel; PK-Merz; Amantadina. IUPAC Name: Tricyclo[3.3.1.13,7]decan-1-amine, hydrochloride Molecular Formula: C10H17NHCl Molecular Weight: 187.71 g/mol Mechanism: Antiparkinsonian activity may be due to inhibition of dopamine reuptake into presynaptic neurons or by increasing dopamine release from presynaptic fibers. Effect: Antiparkinsonian agents; Adamantanes; Dopamine agonists. | Pathogenic genes: PARK2 Mechanistic genes: CCR5, CXCR4, DRD1, DRD2, GRIN3A Metabolic genes: Substrate: COMT, CYP1A2, CYP2B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DDC, UGT1A1, UGT1A9 Transporter genes: SLC22A1 |
 | Name: Apomorphine; 58-00-4; Apomorhin; Apo-go; Apofin; Apokinon; Apokyn; Apomorfina. IUPAC Name: 4H-dibenzo[de,g]quinoline-10,11-diol, 5,6,6a,7-tetrahydro-6-methyl-hydrochloride,hemihydrate. Molecular Formula: C17H17NO2HCl1/2H2O Molecular Weight: 312.79 g/mol Mechanism: Stimulates postsynaptic D2-type receptors within the caudate putamen in the brain. Effect: Antiparkinsonian agents; Non-ergot-derivative dopamine receptor agonists. | Pathogenic genes: PARK2 Mechanistic genes: ADRA2A, ADRA2B, ADRA2C, CALY, DRD1, DRD2, DRD3, DRD4, DRD5, HTR1A, HTR1B, HTR1D, HTR2A, HTR2B, HTR2C Metabolic genes: Substrate: COMT, CYP1A2 (minor), CYP2B6, CYP2C9 (minor), CYP2C19 (minor), CYP2D6, CYP3A4 (minor), CYP3A5, DDC, UGT1A1, UGT1A9 Inhibitor: CYP1A2 (weak), CYP2C19 (weak), CYP3A4 (weak) |
 | Name: Bromocriptine; 25614-03-3; Parlodel; Pravidel; Cycloset; Corpadel; Broman; Bromocriptina. IUPAC Name: Ergotaman-3′-6′-18-trione, 2-bromo-12′-hydroxy-2′-(1-methylethyl)-5′-(2-methylpropyl),monomethanesulfonate,(5′α). Molecular Formula: C32H40BrN5O5CH4SO3 Molecular Weight: 750.70 g/mol Mechanism: Semisynthetic ergot alkaloid derivative and dopamine receptor agonist which activates postsynaptic dopamine receptors in the tuberoinfundibular (inhibiting pituitary prolactin secrection) and nigrostriatal pathways (enhancing coordinated motor control). Causes transient increases in growth hormone secretion in individuals with normal growth hormone concentrations. Paradoxically causes sustained suppression of growth hormone secretion in acromegaly. Dysregulation of brain serotonine activity may also occur. Effect: Antiparkinsonian agents; Ergot-derivative dopamine receptor agonists. | Pathogenic genes: ANKK1, BDNF, GSK3B, LRRK2 Mechanistic genes: ABCB1, AKT1, BDNF, CCK, CCKAR, CCKBR, CNR1, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, GSK3B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2, CY22B6, CYP2C19, CYP2D6, CYP3A4 (major), CYP3A5, DDC, MAOB, UGT1A1, UGT1A9 Inhibitor: CYP1A2 (weak), CYP3A4 (moderate) Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
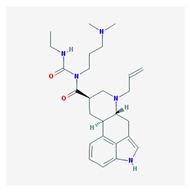 | Name: Cabergoline; 81409-90-7; Cabergoline; Dostinex, Cabaser; Cabergolinum; Cabaseril; Cabergolina. IUPAC Name: Ergoline-8β-carboxamide, N-[3-(dimethylamino)propyl]-N-[(ethylamino)carbonil]-6-(2-propenyl) Molecular Formula: C26H37N5O2 Molecular Weight: 451.60 g/mol Mechanism: A long-acting dopamine receptor agonist. Has high binding affinity for dopamine D2-receptors and lesser affinity for D1, α1- and α2-adrenergic, and serotonin (5-HT1 and 5-HT2) receptors. Reduces serum prolactin concentrations by inhibiting release of prolactin from the anterior pituitary gland (agonist activity at D2 receptors). Effect: Antiparkinsonian agents; Ergot-derivative dopamine receptor agonists. | Pathogenic genes: BDNF, GSK3B Mechanistic genes: ADRA2A, ADRA2B, ADRA2C, AKT1, BDNF, CNR1, DRD1, DRD2, DRD3, DRD4, DRD5, GSK3B, HTR1A, HTR1B, HTR1D, HTR2A, HTR2B, HTR2C, HTR7 Metabolic genes: Substrate: COMT, CYP1A2, CYP2B6, CYP2C19, CYP2D6, CYP3A4 (minor), CYP3A5, DDC |
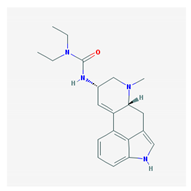 | Name: Lisuride; 18016-80-3; Dopergin; Arolac; Dopergine; Dipergon; Lysenyl; Lisurida. IUPAC Name: 3-(9,10-didehydro-6-methylergolin-8α-yl)-1,1-diethylurea Molecular Formula: C20H26N4O Molecular Weight: 338.45 g/mol Mechanism: Displays dopaminergic, and consequently prolacting-reducing properties. Active substance lisuride has pronounced affinity for dopamine receptors in striatum and pituitary. Effect: Antiparkinsonian agents; Ergot-derivative dopamine receptor agonists. Antimigraine agents. Miscellaneous. | Mechanistic genes: ADRA2A, ADRA2B, ADRA2C, DRD1, DRD2, DRD3, DRD4, DRD5, HTR1A, HTR1B, HTR1D, HTR2A, HTR2B, HTR2C Metabolic genes: Substrate: COMT, CYP1A2, CY22B6, CYP2C19, CYP2D6 (major), CYP3A4 (major), CYP3A5, DDC, UGT1A1, UGT1A9 |
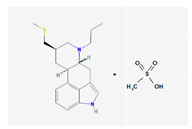 | Name: Pergolide; 66104-22-1; Pergolide; Permax; Pergolida; Pergolidum. IUPAC Name: Ergoline,8-[(methylthio)methyl]-6-monomethenesulfonate Molecular Formula: C19H26N2SCH4O3S Molecular Weight: 410.59 g/mol Mechanism: A dopamin receptor agonist. Relieves symptoms of parkinsonism, presumably by directly stimulating postsynaptic dopamine receptors in corpus striatum. Reduces serum prolactine concentrations by inhibiting release of prolactin from anterior pituitary gland. Causes transient increase in serum somatotropin (growth hormone) concentrations and decreases in serum luteinizing hormone concentrations. Effect: Antiparkinsonian agents; Ergot-derivative dopamine receptor agonists. | Mechanistic genes: ADRA1A, ADRA1B, ADRA1D, ADRA2A, ADRA2B, ADRA2C, DRD1, DRD2, DRD3, DRD4, DRD5, HTR1A, HTR1B, HTR1D, HTR2A, HTR2B, HTR2C Metabolic genes: Substrate: COMT, CYP1A2, CY22B6, CYP2C19, CYP2D6, CYP3A4 (major), CYP3A5, DDC, UGT1A1, UGT1A9 Transporter genes: SLC6A4 |
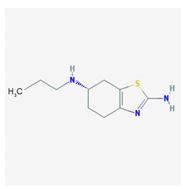 | Name: Pramipexole; 104632-26-0; Pramipexole; Pramipexol; Parmital; Mirapex; Mirapexin; Sifrol IUPAC Name: 2,6-benzothiazolediamine, 4,5,6,7-tetrahydro-N6-propyl-,(S) Molecular Formula: C10H17N3S Molecular Weight: 211.33 g/mol Mechanism: By binding to D2 subfamily dopamine receptor, and to D3, and D4 receptors, it is though that Pramipexole can stimulate dopamine activity on nerves of striatum and substantia nigra. Effect: Antiparkinsonian agents; Non-ergot-derivative dopamine receptor agonists. | Pathogenic genes: ANKK1, BDNF, LRRK2 Mechanistic genes: ADRA2A, ADRA2B, ADRA2C, CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, HTR1A, HTR1B, HTR1D, HTR2A, HTR2B, HTR2C, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2, CY22B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DDC, MAOB, UGT1A1, UGT1A9 Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
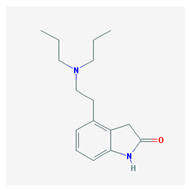 | Name: Ropinirole; 91374-21-9; Ropinirole; ReQuip; Ropinirol; Ropinilorum; ReQuip CR IUPAC Name: 2-H-Indol-2-one 4-[2-(dipropylamino)ethyl]-1,3-dihydro-, monohydrochloride Molecular Formula: C16H24N2O Molecular Weight: 296.84 g/mol Mechanism: Has high relative in vitro specificity and full intrinsic activity at D2 and D3 dopamine receptor subtypes, binding with higher affinity to D3 than to D2 and D4 receptor subtypes. Although precise mechanism of action unknown, it is believed to be due to stimulation of postsynaptic dopamine D2-type receptors within caudate putamen in brain. Mechanism of Ropinirole-induced postural hypotension believed to be due to D2-mediated blunting of noradrenergic response to standing and subsequent decrease in peripheral vascular resistance. Effect: Antiparkinsonian agents; Non-ergot-derivative dopamine receptor agonists. | Pathogenic genes: ANKK1, BDNF, LRRK2 Mechanistic genes: ADRA2A, ADRA2B, ADRA2C, CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, HTR1A, HTR1B, HTR1D, HTR2A, HTR2B, HTR2C, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2 (major), CY22B6, CYP2C19, CYP2D6, CYP3A4 (minor), CYP3A5, DDC, MAOB, UGT1A1, UGT1A9 Inhibitor: CYP1A2 (moderate), CYP2D6 (moderate), CYP3A4 (moderate) Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
 | Name: Rotigotine; 99755-59-6; Rotigotine; Rotigotina; Neupro IUPAC Name: 1-Naphthalenol, 5,6,7,8-tetrahydro-6-[propyl[2-(2-thienyl)ethyl]amino]-6S Molecular Formula: C19H25NOs Molecular Weight: 315.47 g/mol Mechanism: A non-ergot dopamine receptor agonist with specificity for D3-, D2-, and D1-dopamine receptors. Although precise mechanism of action unknown of Rotigotine, it is believed to be due to stimulation of postsynaptic dopamine D2-type auto receptors within substantia nigra in brain, leading to improved dopaminergic transmission in motor areas in basal ganglia, notably caudate nucleus/putamen regions. Effect: Antiparkinsonian agents; Non-ergot-derivative dopamine receptor agonists. | Pathogenic genes: ANKK1, BDNF, LRRK2 Mechanistic genes: CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, MAOB Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
| Monoamine Oxidase B (MOB) Inhibitors | ||
| Drug | Properties | Pharmacogenetics |
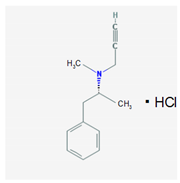 | Name: Selegiline; 14611-51-9; Selegiline; Selegilina; l-Deprenalin; Emsam; Jumex; Eldepryl; Carbex IUPAC Name: Benzeneethanamine,N,α-dimethyl-N-2-propynyl-,hydrochloride,(R) Molecular Formula: C31H17NHCl Molecular Weight: 223.74 g/mol Mechanism: Potent, irreversible inhibitor of the monoamine oxidase (MAO). Plasma concentrations achieved via administration of oral dosage forms in recommended doses confer selective inhibition of the MAO type B, which plays a major role in metabolism of dopamine. Selegiline may also increase dopaminergic activity by interfering with dopamine reuptake at synapse. Effect: Antidepressants. Monoamine oxidase inhibitors. Antiparkinsonian agents. Monoamine oxidase B inhibitors. | Pathogenic genes: ANKK1, BDNF, LRRK2 Mechanistic genes: CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A1, CYP1A2 (minor), CYP1B1, CYP2A6 (minor), CYP2B6 (major), CYP2C8 (minor), CYP2C19 (major), CYP2D6 (minor), CYP2E1 (minor), CYP3A4 (minor), CYP3A5, CYP19A1, DDC, MAOA, MAOB, UGT1A1, UGT1A9 Inhibitor: CYP1A2 (weak), CYP2A6 (weak), CYP2C9 (weak), CYP2C19 (weak), CYP2D6 (weak), CYP2E1 (weak), CYP3A4 (weak), MAOB Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
 | Name: Rasagiline; 136236-51-6; Azilet; Elbrux; Rasagilina; Raxac. IUPAC Name: 1H-Inden-1-amine, 2,3-dihydro-N-2-propynyl-,(R)-, methanesulfonate Molecular Formula: C12H13NCH4O3S Molecular Weight: 267.34 g/mol Mechanism: Potent, irreversible inhibitor of the MAO type B, which plays a major role in catabolism of dopamine. Inhibition of dopamine depletion in striatal region of brain reduces symptomatic motor deficits of Parkinson’s Disease. There is also experimental evidence of Rasagiline conferring neuroprotective effects (antioxidant, antiapoptotic), which may delay onset of symptoms and progression of neuronal deterioration. Effect: Antidepressants. Monoamine oxidase inhibitors. Antiparkinsonian Agents. Monoamine oxidase B inhibitors. | Pathogenic genes: ANKK1, BDNF, LRRK2, PARK2 Mechanistic genes: BLC2, CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2 (major), CYP2B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DDC, MAOB, UGT1A1, UGT1A9 Inhibitor: MAOB Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
| Catecol-O-methyltransferase (COMT) Inhibitors | ||
| Drug | Properties | Pharmacogenetics |
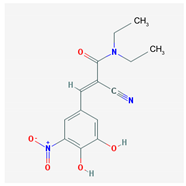 | Name: Entacapone; 130929-57-6; Comtan; Comtess; Entacapona. IUPAC Name: E-α-cyano-N,N-diethyl-3,4-dihydroxy-5-nitrocinnamamida Molecular Formula: C14H15N3O5 Molecular Weight: 305.29 g/mol Mechanism: A selective and selective inhibitor of COMT. When entacapona is taken with levodopa, the pharmacokinetics are altered, resulting in more sustained levodopa serum levels compared to levodopa taken alone. Effect: Antiparkinsonian agents. Catechol-O-methyltransferase inhibitors. | Pathogenic genes: ANKK1, BDNF, LRRK2, PARK2 Mechanistic genes: CCK, CCKAR, CCKBR, DRD1, DRD2, DRD3, DRD4, DRD5, GRIN2A, GRIN2B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2, CYP2B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DDC, MAOB, UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, UGT2B7, UGT2B15 Inhibitor: COMT, CYP1A2 (weak), CYP2A6 (weak), CYP2C9 (weak), CYP2C19 (weak), CYP2D6 (weak), CYP2E1 (weak), CYP3A4 (weak) Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, ACHE, APOE |
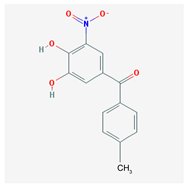 | Name: Tolcapone; 134308-13-7; Tolcapona; Tasmar. IUPAC Name: Methanone, (3,4-hydroxy-5-nitrophenyl)(4-methylphenyl) Molecular Formula: C14H11NO5 Molecular Weight: 273.24 g/mol Mechanism: A selective and selective inhibitor of (COMT. In the presence of a decarboxylase inhibitor (e.g., carbidopa), COMT is the major degradation pathway for levodopa. Inhibition of COMT leads to more sustained plasma levels of levodopa and enhanced central dopaminergic activity. Effect: Antiparkinsonian agents. Catechol-O-methyltransferase inhibitors. | Pathogenic genes: ANKK1, BDNF, LRRK2, PARK2 Mechanistic genes: AKT1, CCK, CCKAR, CCKBR, CNR1, DRD1, DRD2, DRD3, DRD4, DRD5, GPT, GRIN2A, GRIN2B, GSK3B, HCRT, HOMER1, LMO3, OPRM1 Metabolic genes: Substrate: COMT, CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DDC, MAOB, UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, UGT2B7, UGT2B15 Transporter genes: SLC22A1, SLC6A3 Pleiotropic genes: ACE, APOE |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cacabelos, R. Parkinson’s Disease: From Pathogenesis to Pharmacogenomics. Int. J. Mol. Sci. 2017, 18, 551. https://doi.org/10.3390/ijms18030551
Cacabelos R. Parkinson’s Disease: From Pathogenesis to Pharmacogenomics. International Journal of Molecular Sciences. 2017; 18(3):551. https://doi.org/10.3390/ijms18030551
Chicago/Turabian StyleCacabelos, Ramón. 2017. "Parkinson’s Disease: From Pathogenesis to Pharmacogenomics" International Journal of Molecular Sciences 18, no. 3: 551. https://doi.org/10.3390/ijms18030551






