Characterization of Paraquat-Induced miRNA Profiling Response in hNPCs Undergoing Proliferation
Abstract
:1. Introduction
2. Results and Discussion
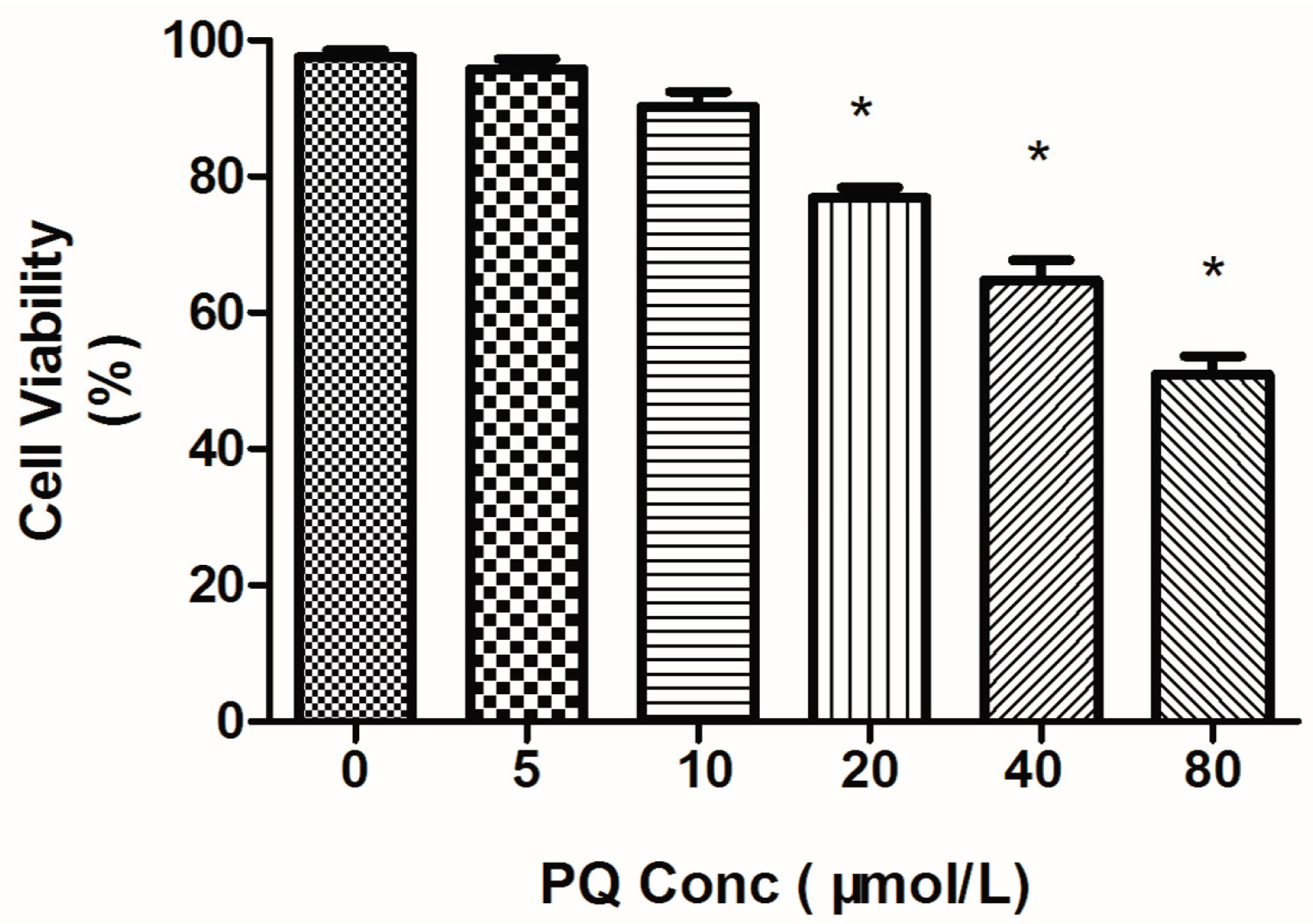
2.1. Effects of Paraquat (PQ) on Human Neural Progenitor Cells (hNPCs) Viability
2.2. Effects of PQ on hNPCs Apoptosis

2.3. Effects of PQ on miRNA Profiling of hNPCs
| microRNA | Fold Change | microRNA | Fold Change |
|---|---|---|---|
| Up-Regulated | Down-Regulated | ||
| hsa-miR-550a-5p | 21.36 | hsa-miR-3182 | 0.01 |
| hsa-miR-550b-2-5p | 13.81 | hsa-miR-4308 | 0.11 |
| hsa-miR-378b | 7.92 | hsa-miR-3679-3p | 0.11 |
| hsa-miR-4784 | 6.50 | hsa-miR-145-3p | 0.12 |
| hsa-miR-3142 | 5.30 | hsa-miR-1273g-3p | 0.15 |
| hsa-let-7f-5p | 5.19 | hsa-miR-124-3p | 0.18 |
| hsa-miR-3175 | 4.99 | hsa-miR-3178 | 0.27 |
| hsa-miR-4446-5p | 4.29 | hsa-miR-3675-3p | 0.27 |
| hsa-miR-4657 | 3.91 | hsa-miR-1184 | 0.30 |
| hsa-miR-4291 | 3.89 | hsa-miR-299-5p | 0.31 |
| hsa-miR-1304-5p | 3.70 | hsa-miR-491-3p | 0.32 |
| hsa-miR-378d | 3.35 | hsa-miR-4456 | 0.32 |
| hsa-miR-4275 | 3.27 | hsa-miR-208a-5p | 0.33 |
| hsa-miR-10a-5p | 3.26 | hsa-miR-574-3p | 0.33 |
| hsa-miR-195-5p | 3.19 | hsa-miR-589-5p | 0.34 |
| hsa-miR-4289 | 3.12 | hsa-miR-5196-3p | 0.37 |
| hsa-let-7d-5p | 3.06 | hsa-miR-371b-3p | 0.39 |
| hsa-miR-412-3p | 2.99 | hsa-miR-3646 | 0.39 |
| hsa-miR-34c-5p | 2.86 | hsa-miR-1284 | 0.42 |
| hsa-miR-4484 | 2.84 | hsa-miR-1321 | 0.44 |
| hsa-miR-98-5p | 2.72 | hsa-miR-4524b-5p | 0.44 |
| hsa-miR-320d | 2.67 | hsa-miR-4521 | 0.45 |
| hsa-miR-5586-5p | 2.56 | hsa-miR-4305 | 0.45 |
| hsa-miR-27a-3p | 2.43 | hsa-miR-498 | 0.46 |
| hsa-miR-3654 | 2.40 | hsa-miR-944 | 0.47 |
| hsa-miR-3686 | 2.39 | hsa-miR-519e-5p | 0.48 |
| hsa-miR-3135a | 2.37 | ||
| hsa-miR-378e | 2.32 | ||
| hsa-miR-320e | 2.30 | ||
| hsa-let-7g-5p | 2.23 | ||
| hsa-miR-30c-5p | 2.19 | ||
| hsa-miR-1290 | 2.17 | ||
| hsa-miR-30c-1-3p | 2.16 | ||
| hsa-miR-484 | 2.15 | ||
| hsa-miR-23a-3p | 2.10 | ||
| hsa-miR-647 | 2.09 | ||
| hsa-miR-4742-3p | 2.08 | ||
| hsa-miR-23c | 2.05 | ||
| hsa-miR-767-5p | 2.05 | ||
| hsa-miR-376a-3p | 2.04 | ||
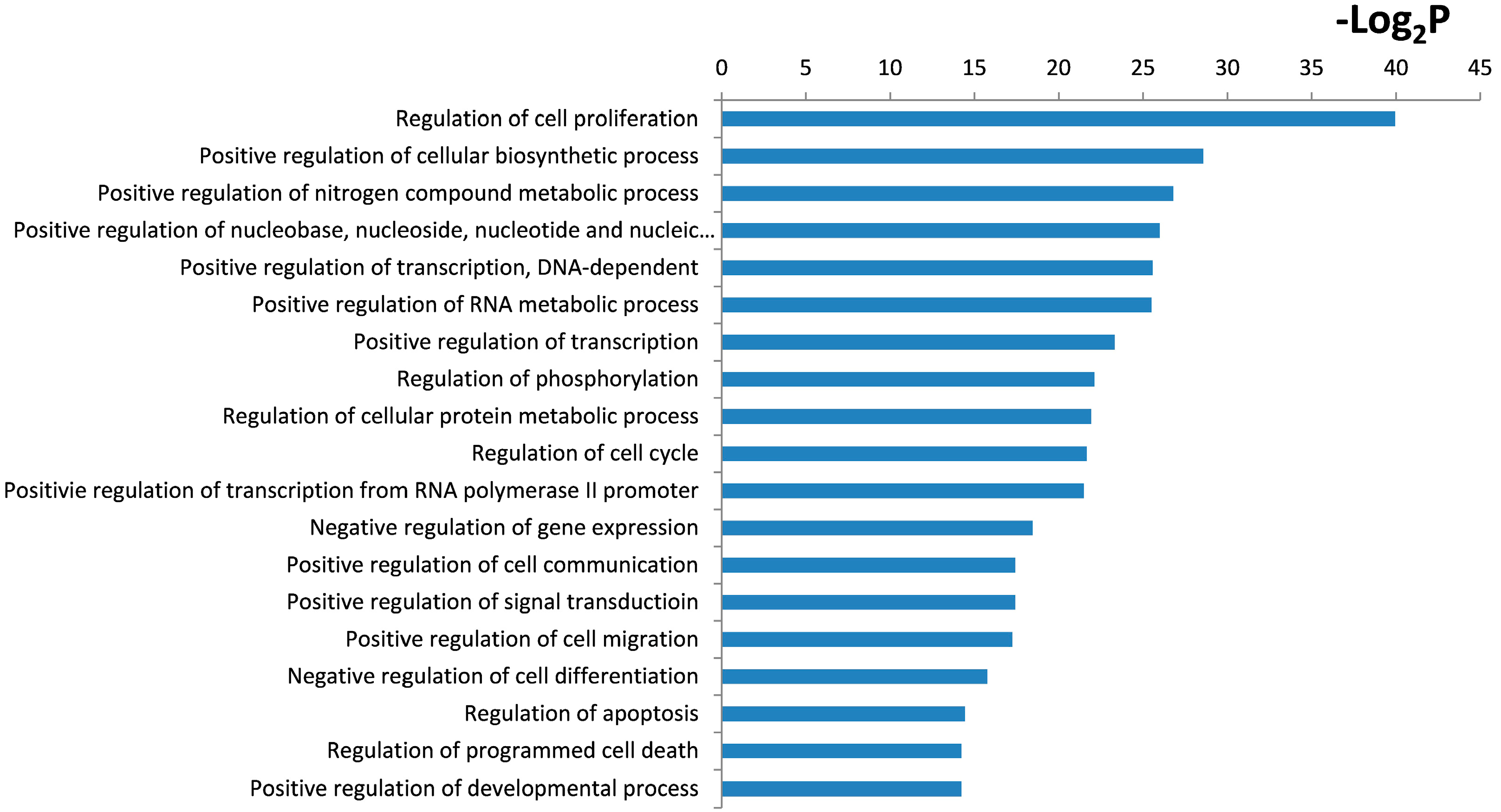
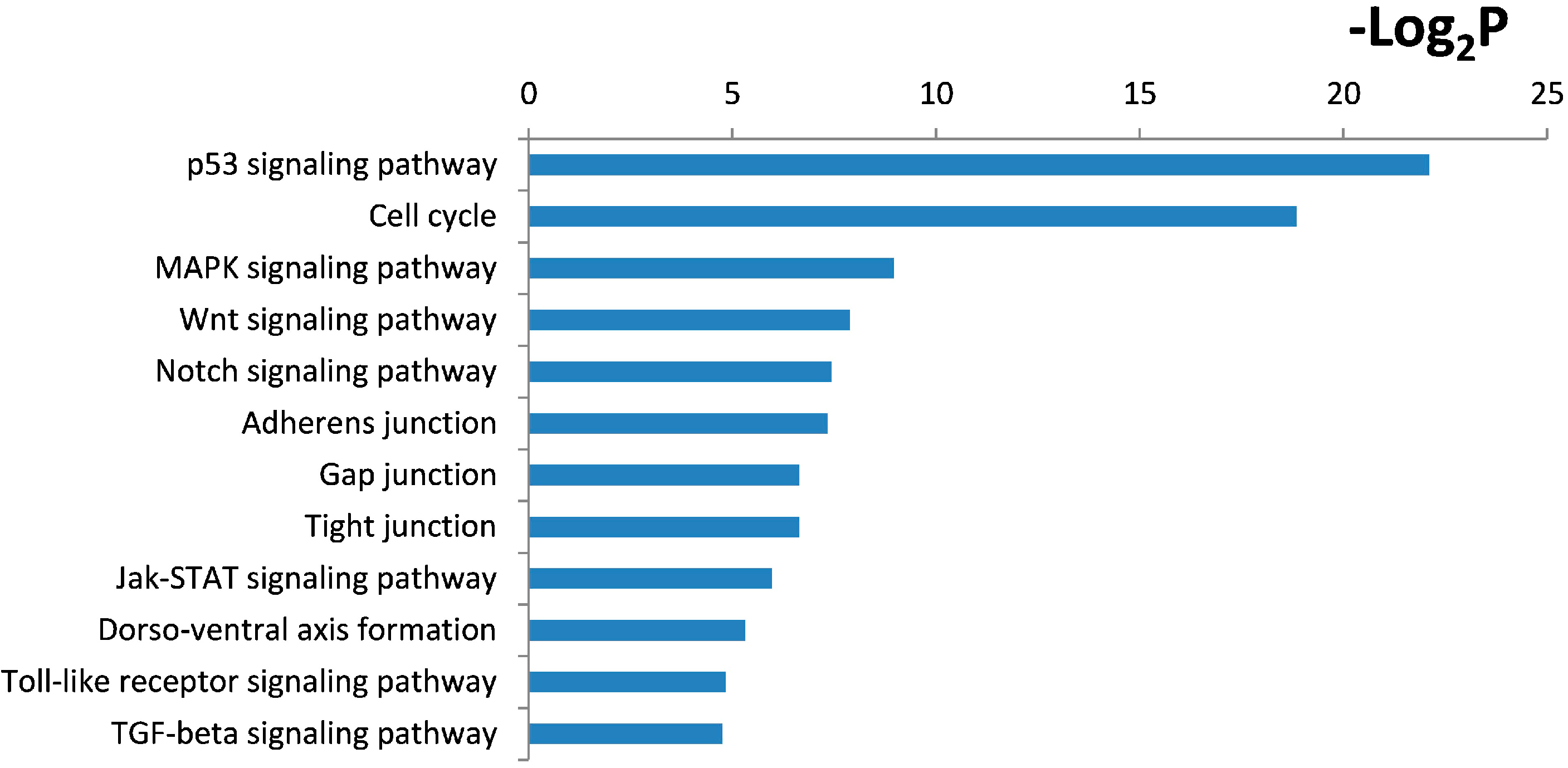
2.4. Functional and Pathway Analysis
2.5. Real-Time RT-PCR Validation of PQ-Responsive miRNAs and Target Genes
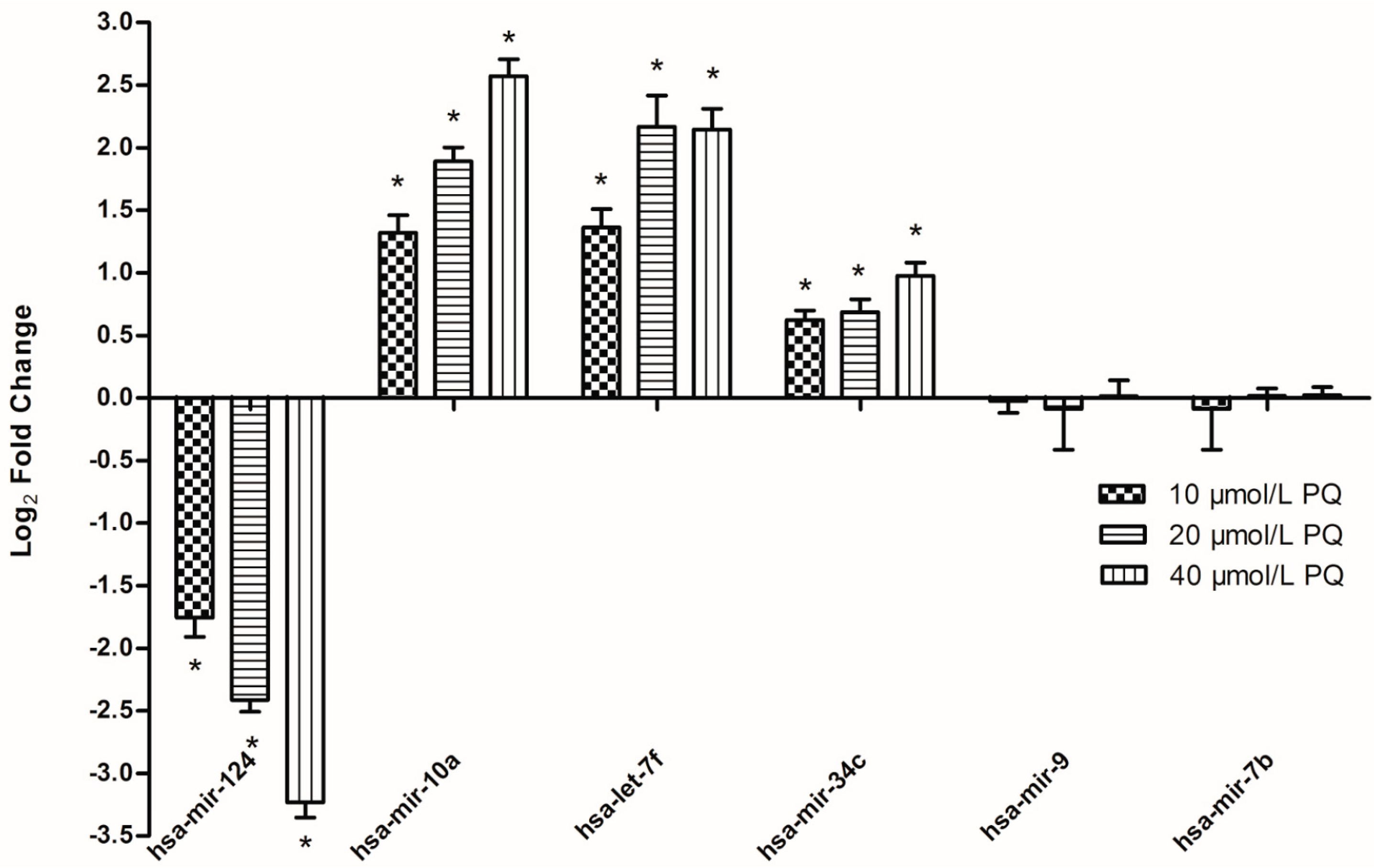
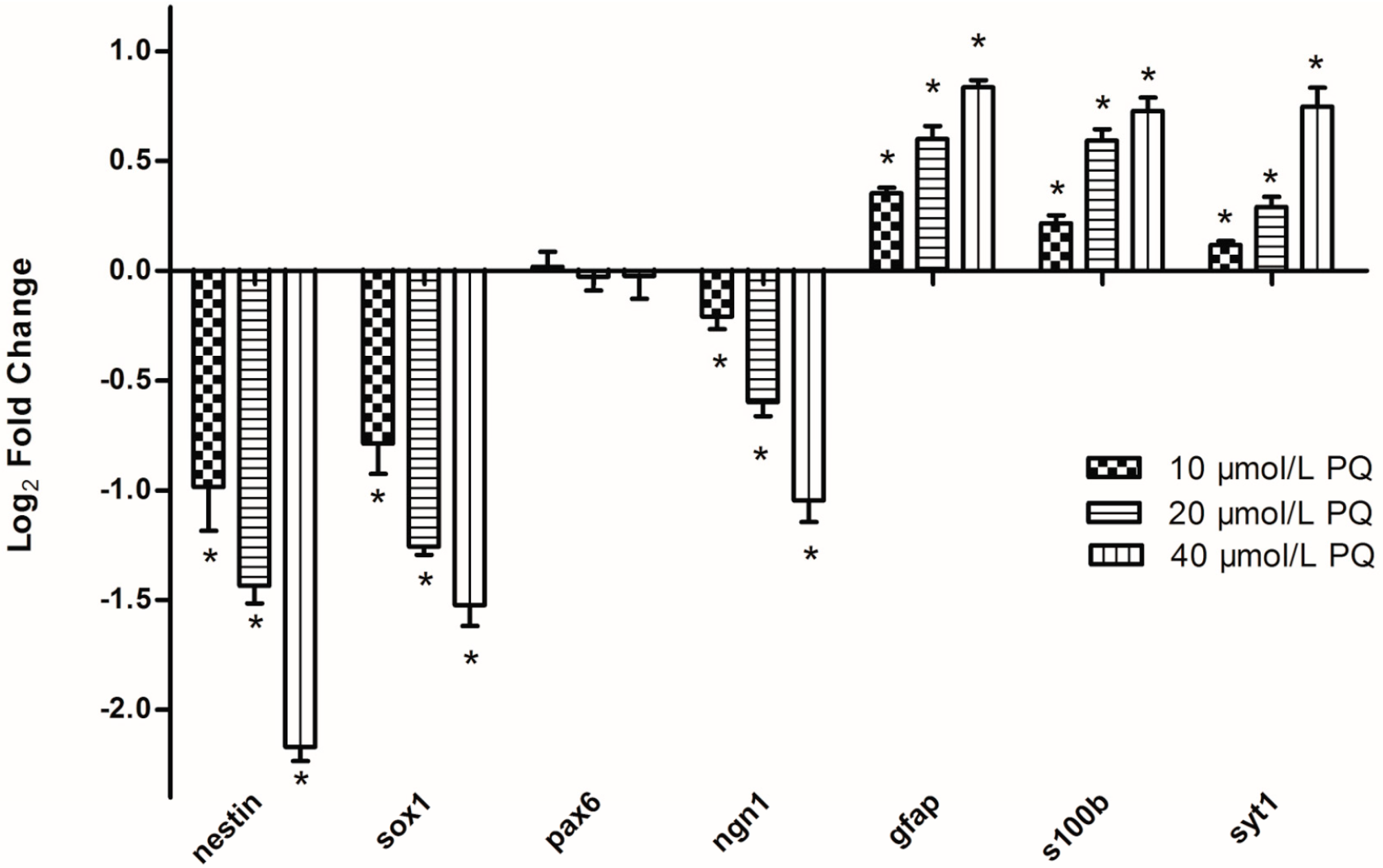
2.6. Discussion
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Cell Culture and Treatments
3.3. Cell Viability Assay
3.4. Cell Apoptosis Assay
3.5. RNA Isolation
3.6. miRNA Microarray Analysis
3.7. Target Prediction and Functional Analysis
3.8. Real-Time RT-PCR Validation
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Prasad, K.; Tarasewicz, E.; Mathew, J.; Strickland, P.A.O.; Buckley, B.; Richardson, J.R.; Richfield, E.K. Toxicokinetics and toxicodynamics of paraquat accumulation in mouse brain. Exp. Neurol. 2009, 215, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.; Chang, X.; Li, W.; Zhao, Q.; Wang, Y.; Zhou, A. Paraquat affects the homeostasis of dopaminergic system in PC12 cells. Pestic. Biochem. Physiol. 2012, 103, 81–86. [Google Scholar] [CrossRef]
- Djukic, M.M.; Jovanovic, M.D.; Ninkovic, M.; Stevanovic, I.; Ilic, K.; Curcic, M.; Vekic, J. Protective role of glutathione reductase in paraquat induced neurotoxicity. Chem. Biol. Interact. 2012, 199, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Rappold, P.M.; Cui, M.; Chesser, A.S.; Tibbett, J.; Grima, J.C.; Duan, L.; Sen, N.; Javitch, J.A.; Tizeu, K. Paraquat neurotoxicity is mediated by the dopamine transporter and organic cation transporter-3. Proc. Natl. Acad. Sci. USA 2011, 108, 20766–20771. [Google Scholar] [CrossRef] [PubMed]
- Tsatsakis, A.M.; Perakis, K.; Koumantakis, E. Experience with acute paraquat poisoning in Crete. Vet. Hum. Toxicol. 1996, 38, 113–117. [Google Scholar]
- Grandjean, P.; Landrigan, P.J. Neurobehavioural effects of developmental toxicity. Lancet Neurol. 2014, 13, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, Y.; Gotoh, Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 2010, 11, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Meza-Sosa, K.F.; Pedraza-Alva, G.; Perez-Martinez, L. microRNAs: Key triggers of neuronal cell fate. Front. Cell. Neurosci. 2014, 8, 175. [Google Scholar] [CrossRef] [PubMed]
- Kintner, C. Neurogenesis in embryos and in adult neural stem cells. J. Neurosci. 2002, 22, 639–643. [Google Scholar] [PubMed]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk-database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, P. Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 2010, 11, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Block, K.; Sittka, A.; Oelgeschlager, M.; Seiler, A.E.M.; Luch, A. MicroRNA profiling as tool for in vitro developmental neurotoxicity testing: The case of sodium valproate. PLoS One 2014, 9, e98892. [Google Scholar] [CrossRef]
- Chang, X.; Lu, W.; Dou, T.; Wang, X.; Lou, D.; Sun, X.; Zhou, Z. Paraquat inhibits cell viability via enhanced oxidative stress and apoptosis in human neural progenitor cells. Chem. Biol. Interact. 2013, 206, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef] [PubMed]
- Noorbakhsh, F.; Ellestad, K.K.; Maingat, F.; Warren, K.G.; Han, M.H.; Steinman, L.; Baker, G.B.; Power, C. Impaired neurosteroid synthesis in multiple sclerosis. Brain 2011, 134, 2703–2721. [Google Scholar] [CrossRef] [PubMed]
- Barbeau, A.; Roy, M.; Cloutier, T.; Plasse, L.; Paris, S. Environmental and genetic factors in the etiology of Parkinson’s disease. Adv. Neurol. 1987, 45, 299–306. [Google Scholar] [PubMed]
- Liou, H.H.; Tsai, M.C.; Chen, C.J.; Jeng, J.S.; Chang, Y.C.; Chen, S.Y.; Chen, R.C. Environmental risk factors and Parkinson’s disease: A case-control study in Taiwan. Neurology 1997, 48, 1583–1588. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.; Lv, X.; Jiao, J. The role of microRNAs in neural stem cells and neurogenesis. J. Genet. Genomics 2013, 40, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.Y.; Chung, K.H.; Deo, M.; Thompson, R.C.; Turner, D.L. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 2008, 314, 2618–2633. [Google Scholar] [CrossRef] [PubMed]
- Delaloy, C.; Liu, L.; Lee, J.A.; Su, H.; Shen, F.; Yang, G.Y.; Young, W.L.; Ivey, K.N.; Gao, F.B. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 2010, 6, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Wulczyn, F.G.; Smirnova, L.; Rybak, A.; Brandt, C.; Kwidzinski, E.; Ninnemann, O.; Strehle, M.; Seiler, A.; Schumacher, S.; Nitsch, R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007, 21, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, G.; Li, S.; Lang, M.; Yang, S.; Li, W.; Shi, Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Davoodian, N.; Lotfi, A.S.; Soleimani, M.; Mola, S.J.; Arjmand, S. Let-7f microRNA negatively regulates hepatic differentiation of human adipose tissue-derived stem cells. J. Physiol. Biochem. 2014, 70, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Redline, R.W.; Neish, A.; Holmes, L.B.; Collins, T. Homeobox genes and congenital malformations. Lab. Investig. 1992, 66, 659–670. [Google Scholar] [PubMed]
- Meseguer, S.; Mudduluru, G.; Escamilla, J.M.; Allgayer, H.; Barettino, D. MicroRNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF). J. Biol. Chem. 2011, 286, 4150–4164. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Medina, R.; Cayuso, J.; Okubo, T.; Takada, S.; Marti, E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development 2008, 135, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.D.; Tseng, Y.T.; Shrestha, S.; Lin, Y.L.; Khaleel, A.; Chou, C.H.; Chu, C.F.; Huang, H.Y.; Lin, C.M.; Ho, S.Y.; et al. miRTarBase update 2014: An information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014, 42, D78–D85. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Lou, D.; Cai, Q.; Chang, X.; Wang, X.; Zhou, Z. Characterization of Paraquat-Induced miRNA Profiling Response in hNPCs Undergoing Proliferation. Int. J. Mol. Sci. 2014, 15, 18422-18436. https://doi.org/10.3390/ijms151018422
Huang M, Lou D, Cai Q, Chang X, Wang X, Zhou Z. Characterization of Paraquat-Induced miRNA Profiling Response in hNPCs Undergoing Proliferation. International Journal of Molecular Sciences. 2014; 15(10):18422-18436. https://doi.org/10.3390/ijms151018422
Chicago/Turabian StyleHuang, Min, Dan Lou, Qian Cai, Xiuli Chang, Xinjin Wang, and Zhijun Zhou. 2014. "Characterization of Paraquat-Induced miRNA Profiling Response in hNPCs Undergoing Proliferation" International Journal of Molecular Sciences 15, no. 10: 18422-18436. https://doi.org/10.3390/ijms151018422




