Novel Y Chromosome Retrocopies in Canids Revealed through a Genome-Wide Association Study for Sex
Abstract
1. Introduction
2. Materials and Methods
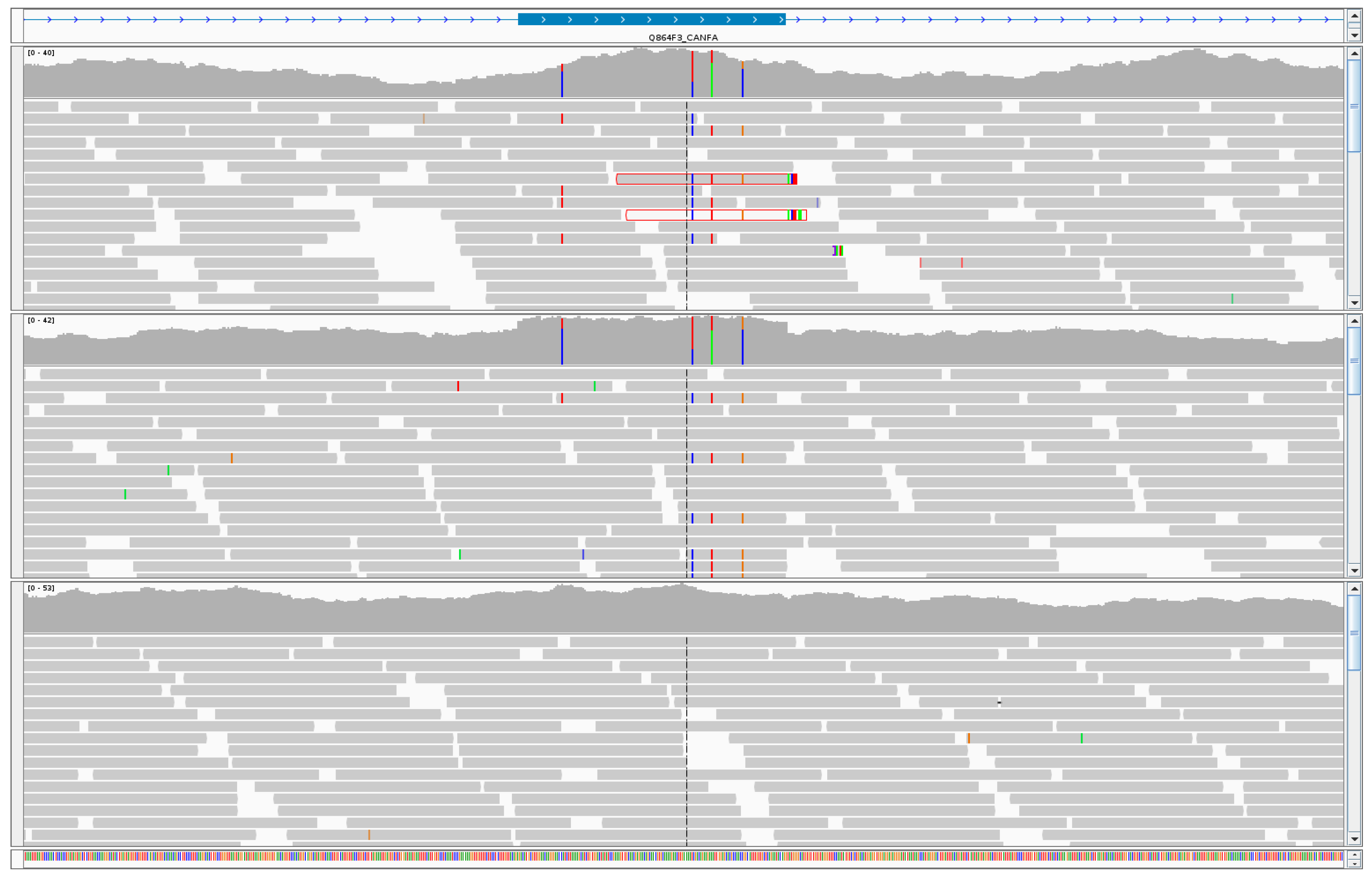
3. Results
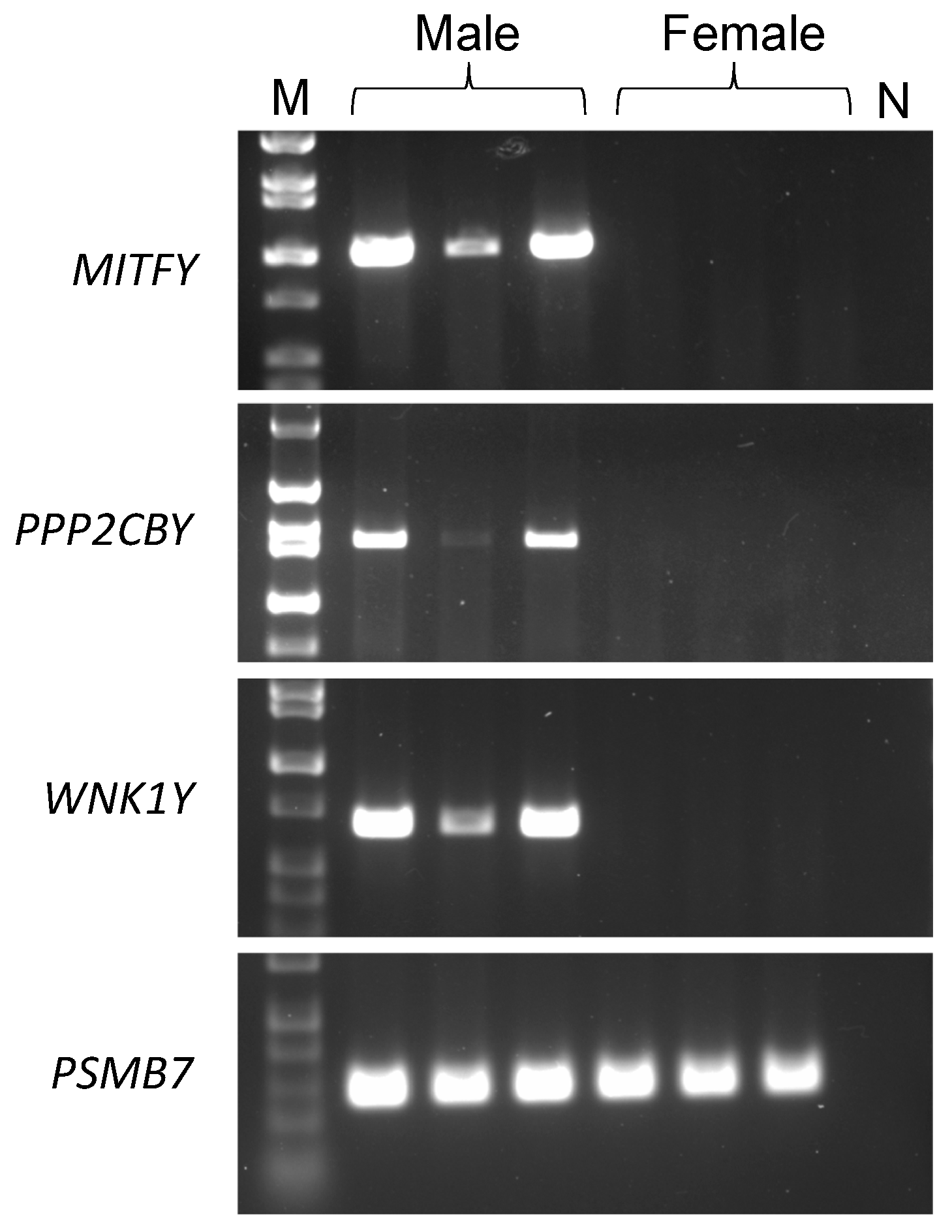
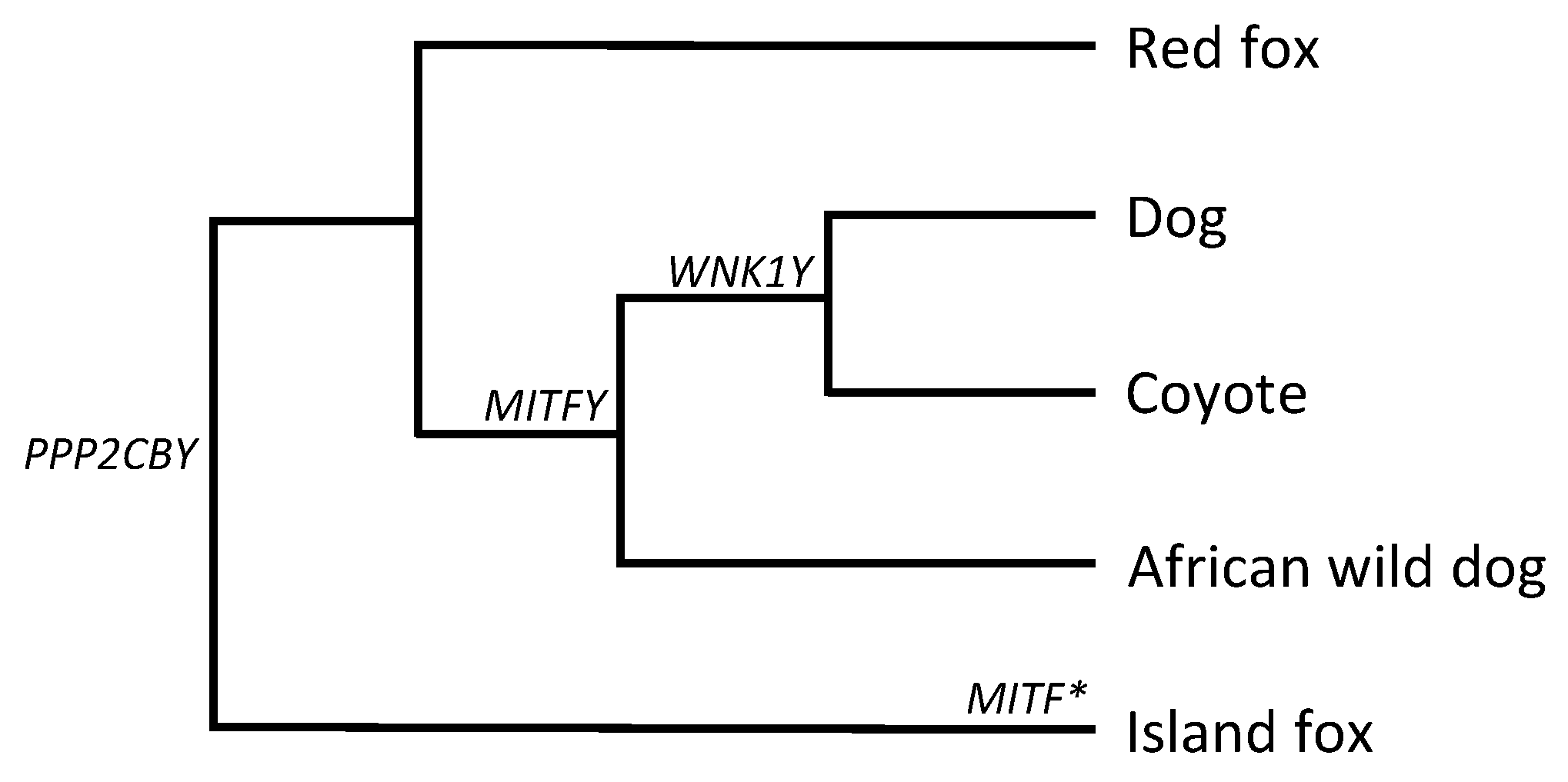
3.1. MITFY
3.2. PPP2CBY
3.3. WNK1Y
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef]
- Yang, Y.; Chang, T.C.; Yasue, H.; Bharti, A.K.; Retzel, E.F.; Liu, W.S. ZNF280BY and ZNF280AY: autosome derived Y-chromosome gene families in Bovidae. BMC Genomics 2011, 13. [Google Scholar] [CrossRef]
- Dyer, K.A.; White, B.E.; Bray, M.J.; Piqué, D.G.; Betancourt, A.J. Molecular evolution of a Y chromosome to autosome gene duplication in Drosophila. Mol. Biol. Evol. 2011, 28, 1293–1306. [Google Scholar] [CrossRef][Green Version]
- Hughes, J.F.; Skaletsky, H.; Koutseva, N.; Pyntikova, T.; Page, D.C. Sex chromosome to autosome transposition events counter Y-chromosome gene loss in mammals. Genome Biol. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Page, D.C.; Harper, M.E.; Love, J.; Botstein, D. Occurence of a transposition from the X-chromosome long arm to the Y-chromosome short arm during human evolution. Nature 1984, 311, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Brown, L.G.; Hawkins, T.; Alagappan, R.K.; Skaletsky, H.; Reeve, M.P.; Reijo, R.; Rozen, S.; Dinulos, M.B.; Disteche, C.M.; Page, D.C. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996, 14, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Lahn, B.T.; Page, D.D. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat. Genet. 1999, 21, 429–433. [Google Scholar] [CrossRef]
- Cao, P.; Wang, L.; Jiang, Y.; Yi, Y.; Qu, F.; Liu, T.; Lv, L. De Novo origin of VCY2 from autosome to y transposed amplicon. PLoS ONE 2015, 10, e0119651. [Google Scholar] [CrossRef][Green Version]
- Carvalho, A.B.; Vicoso, B.; Russo, C.A.M.; Swenor, B.; Clark, A.G. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2015, 112, 12450–12455. [Google Scholar] [CrossRef]
- Marques, A.C.; Dupanloup, I.; Vinckenbosch, N.; Reymond, A.; Kaessmann, H. Emergence of young human genes after a burst of retroposition in primates. PLoS ONE 2005, 3, e357. [Google Scholar] [CrossRef]
- Bai, Y.; Casola, C.; Betrán, E. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 2008, 9, 241. [Google Scholar] [CrossRef] [PubMed]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J., III; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Hoeppner, M.P.; Lundquist, A.; Pirun, M.; Meadows, J.R.S.; Zamani, N.; Johnson, J.; Sundström, G.; Cook, A.; FitzGerald, M.G.; Swofford, R.; Mauceli, E.; et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE 2014, 9, e91172. [Google Scholar] [CrossRef] [PubMed]
- Natanaelsson, C.; Oskarsson, M.C.R.; Angleby, H.; Lundeberg, J.; Kirkness, E.; Savolainen, P. Dog Y chromosomal DNA sequence: identification, sequencing and SNP discovery. BMC Genet. 2006, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Davis, B.W.; Raudsepp, T.; Pearks Wilkerson, A.J.; Mason, V.C.; Ferguson-Smith, M.; O’Brien, P.C.; Waters, P.D.; Murphy, W.J. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 2013, 23, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 7, 873–881. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Parker, H.G.; VonHoldt, B.M.; Quignon, P.; Margulies, E.H.; Shao, S.; Mosher, D.S.; Spady, T.C.; Elkahloun, A.; Cargill, M.; Jones, P.G.; et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 2009, 325, 995–998. [Google Scholar] [CrossRef]
- Brown, E.A.; Dickinson, P.J.; Mansour, T.; Sturges, B.K.; Aguilar, M.; Young, A.E.; Korff, C.; Lind, J.; Ettinger, C.L.; Varon, S.; et al. FGF4 retrogene on CFA12 is responsible for chondrodystrophy and intervertebral disc disease in dogs. Proc. Natl. Acad. Sci. USA 2017, 114, 11476–11481. [Google Scholar] [CrossRef] [PubMed]
- Marchant, T.W.; Johnson, E.J.; McTeir, L.; Johnson, C.I.; Gow, A.; Liuti, T.; Kuehn, D.; Svenson, K.; Bermingham, M.L.; Drögemüller, M.; et al. Canine Brachycephaly is associated with a retrotransposon-mediated missplicing of SMOC2. Curr. Biol. 2017, 27, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Morais, D.; Ivanga, M.; Harrison, P.M. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinformatics 2007, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Potrzebowski, L.; Vinckenbosch, N.; Marques, A.C.; Chalmel, F.; Jégou, B.; Kaessmann, H. Chromosomal gene movement reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008, 6, e80. [Google Scholar] [CrossRef]
- Pan, D.; Zhang, L. Burst of young retrogenes and independent retrogene formation in mammals. PLoS ONE 2009, 4, e5040. [Google Scholar] [CrossRef] [PubMed]
- Fablet, M.; Bueno, M.; Potrzebowski, L.; Kaessmann, H. Evolutionary origin and function of retrogene introns. Mol. Biol. Evol. 2009, 26, 2147–2156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lindeboom, R.G.; Supek, F.; Lehner, B. The rules and impact of nonsense mediated RNA decay in human cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef]
- Hemesath, T.J.; Steingrímsson, E.; McGill, G.; Hansen, M.J.; Vaught, J.; Hodgkinson, C.A.; Arnheiter, H.; Copeland, N.G.; Jenkins, N.A.; Fisher, D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes. Dev. 1994, 8, 2770–2780. [Google Scholar] [CrossRef]
- Saito, H.; Takeda, K.; Yasumoto, K.; Ohtani, H.; Watanabe, K.; Takahashi, K.; Fukuzaki, A.; Arai, Y.; Yamamoto, H.; Shibahara, S. Germ cell-specific expression of microphthalmia associated transcription factor mRNA in mouse testis. J. Biochem. 2003, 134, 143–150. [Google Scholar] [CrossRef]
- Hartman, M.L.; Czyz, M. MITF in melanoma: mechanisms behind its expression and activity. Cell. Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef]
- Vidal-Petiot, E.; Cheval, L.; Faugeroux, J.; Malard, T.; Doucet, A.; Jeunemaitre, X.; Hadchouel, J. A new methodology for quantification of alternatively spliced exons reveals a highly tissue-specific expression pattern of WNK1 isoforms. PLoS ONE 2012, 7, e37751. [Google Scholar] [CrossRef]
- Eo, J.; Song, H.; Lim, H.J. Etv5, a transcription factor with versatile functions in male reproduction. Clin. Exp. Reprod. Med. 2012, 39, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Shin, S.; Janknecht, R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta 2012, 1826, 1–12. [Google Scholar] [CrossRef]
- Bai, Y.; Casola, C.; Feschotte, C.; Betrán, E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007, 8, R11. [Google Scholar] [CrossRef]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Rosikiewicz, W.; Kabza, M.; Kosiński, J.G.; Ciomborowska-Basheer, J.; Kubiak, M.R.; Makałowska, I. Retrogene DB — a database of plant and animal retrocopies. Database 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- O’Bryan, M.K.; Grealy, A.; Stahl, P.J.; Schlegel, P.N.; McLachlan, R.I.; Jamsai, D. Genetic variants in the ETV5 gene in fertile and infertile men with nonobstructive azoospermia associated with Sertoli cell-only syndrome. Fertil. Steril. 2012, 98, 827–835. [Google Scholar] [CrossRef]
- Su, Y.; Sugiura, K.; Sun, F.; Pendola, J.K.; Cox, G.A.; Handel, M.A.; Schimenti, J.C.; Eppig, J.J. MARF1 regulates essential oogenic processes in mice. Science 2012, 335, 1496–1499. [Google Scholar] [CrossRef]
- Shekarabi, M.; Lafrenière, R.G.; Gaudet, R.; Laganière, J.; Marcinkiewicz, M.M.; Dion, P.A.; Rouleau, G.A. Comparative analysis of the expression profile of Wnk1 and Wnk1/Hsn2 splice variants in developing and adult mouse tissues. PLoS ONE 2013, 8, e57807. [Google Scholar] [CrossRef] [PubMed]
- Rohozinski, J. Lineage-independent retrotransposition of UTP14 associated with male fertility has occurred multiple times throughout mammalian evolution. R. Soc. Open Sci. 2017, 4, 171049. [Google Scholar] [CrossRef] [PubMed]
- Ciomborowska, J.; Rosikiewicz, W.; Szklarczyk, D.; Makałowski, W.; Makałowska, I. “Orphan” retrogenes in the human genome. Mol. Biol. Evol. 2012, 30, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Skinner, B.M.; Sargent, C.A.; Churcher, C.; Hunt, T.; Herrero, J.; Loveland, J.E.; Dunn, M.; Louzada, S.; Fu, B.; Chow, W. The pig X and Y chromosomes: structure, sequence, and evolution. Genome Res. 2016, 26, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Klabnik, J.L.; Liu, W.S. Regional selection acting on the OFD1 gene family. PLoS ONE 2011, 6, e26195. [Google Scholar] [CrossRef] [PubMed]
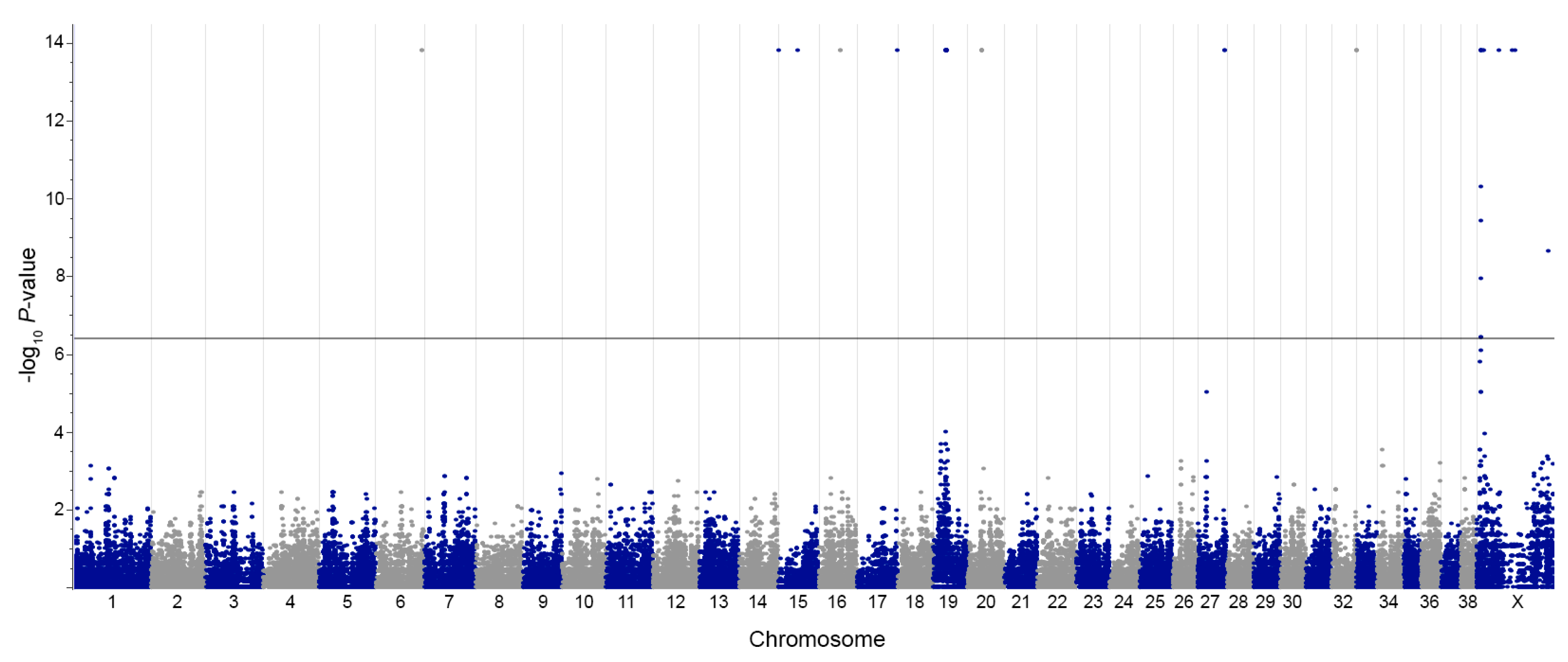

| Chr | Position | Gene | Region | Chr | Position | Gene | Region |
|---|---|---|---|---|---|---|---|
| 6 | 72952463 | 20 | 21870155 | MITF | exonic | ||
| 72964379 | 21870230 | MITF | exonic | ||||
| 15 | 111194 | 21870310 | MITF | exonic | |||
| 29973349 | 21870623 | MITF | 3′ UTR | ||||
| 16 | 33566554 | PPP2CB | 3′ UTR | 21871904 | MITF | 3′ UTR | |
| 33566935 | PPP2CB | 3′ UTR | 21872335 | MITF | 3′ UTR | ||
| 17 | 64209287 | 21872815 | MITF | 3′ UTR | |||
| 19 | 20034966 | 21873532 | MITF | 3′ UTR | |||
| 20040153 | 27 | 42911747 | WNK1 | exonic | |||
| 20052361 | 42911917 | WNK1 | exonic | ||||
| 20062503 | 32 | 38734101 | |||||
| 20072295 | 38767343 | ||||||
| 20100030 | 38788489 | ||||||
| 20114792 | 38789367 | ||||||
| 20130159 | X | 6604781 | SHROOM2 | exonic | |||
| 20152245 | 6621021 | SHROOM2 | intronic | ||||
| 20172164 | 6628533 | SHROOM2 | intronic | ||||
| 20245553 | 6634742 | SHROOM2 | intronic | ||||
| 20256174 | 10131021 | TRAPPC2 | exonic | ||||
| 20286580 | 10175834 | OFD1 | exonic | ||||
| 20292945 | 35604689 | USP9X/Y | exonic | ||||
| 20303661 | 57139861 | ||||||
| 20309777 | 60395963 | PGK1 | |||||
| 20310190 | Y | 26641 | |||||
| 20314276 | 316950 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, K.L.; Evans, J.M.; Noorai, R.E.; Starr-Moss, A.N.; Clark, L.A. Novel Y Chromosome Retrocopies in Canids Revealed through a Genome-Wide Association Study for Sex. Genes 2019, 10, 320. https://doi.org/10.3390/genes10040320
Tsai KL, Evans JM, Noorai RE, Starr-Moss AN, Clark LA. Novel Y Chromosome Retrocopies in Canids Revealed through a Genome-Wide Association Study for Sex. Genes. 2019; 10(4):320. https://doi.org/10.3390/genes10040320
Chicago/Turabian StyleTsai, Kate L., Jacquelyn M. Evans, Rooksana E. Noorai, Alison N. Starr-Moss, and Leigh Anne Clark. 2019. "Novel Y Chromosome Retrocopies in Canids Revealed through a Genome-Wide Association Study for Sex" Genes 10, no. 4: 320. https://doi.org/10.3390/genes10040320
APA StyleTsai, K. L., Evans, J. M., Noorai, R. E., Starr-Moss, A. N., & Clark, L. A. (2019). Novel Y Chromosome Retrocopies in Canids Revealed through a Genome-Wide Association Study for Sex. Genes, 10(4), 320. https://doi.org/10.3390/genes10040320





