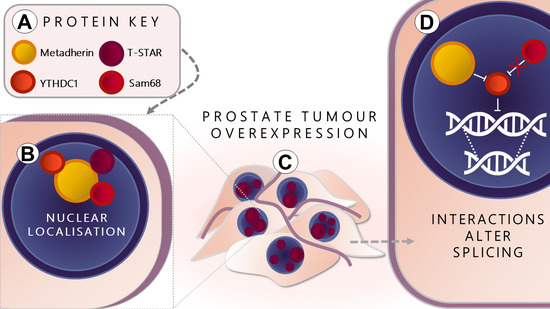
The Oncogene Metadherin Interacts with the Known Splicing Proteins YTHDC1, Sam68 and T-STAR and Plays a Novel Role in Alternative mRNA Splicing
Abstract
:1. Introduction
2. Results
2.1. Metadherin Interacts with Splicing Protein Ythdc1
2.2. Sam68-Associated Protein T-Star is Androgen-Regulated and Overexpressed in Prostate Cancer
2.3. Metadherin Directly Interacts with Splicing Complex Proteins Sam68 and T-Star
2.4. Metadherin Attenuates the Effect of Ythdc1 on Cd44v5-Luc Splicing
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmids
4.3. CD44 Minigene Assays
4.4. Yeast Two-Hybrid Assay
4.5. Cell lysates, Western Blotting and Immunoprecipitation
4.6. Confocal Microscopy
4.7. Tissue Microarray and Immunohistochemistry
4.8. Statistics and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hsu, J.C.; Reid, D.W.; Hoffman, A.M.; Sarkar, D.; Nicchitta, C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 2018, 24, 688–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emdad, L.; Das, S.K.; Hu, B.; Kegelman, T.; Kang, D.C.; Lee, S.G.; Sarkar, D.; Fisher, P.B. AEG-1/MTDH/LYRIC: A Promiscuous Protein Partner Critical in Cancer, Obesity, and CNS Diseases. Adv. Cancer Res. 2016, 131, 97–132. [Google Scholar] [PubMed]
- Li, J.; Yang, L.; Song, L.; Xiong, H.; Wang, L.; Yan, X.; Yuan, J.; Wu, J.; Li, M. Astrocyte elevated gene-1 is a proliferation promoter in breast cancer via suppressing transcriptional factor FOXO1. Oncogene 2009, 28, 3188–3196. [Google Scholar] [CrossRef] [PubMed]
- Kikuno, N.; Shiina, H.; Urakami, S.; Kawamoto, K.; Hirata, H.; Tanaka, Y.; Place, R.F.; Pookot, D.; Majid, S.; Igawa, M.; et al. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene 2007, 26, 7647–7655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirkettle, H.J.; Mills, I.G.; Whitaker, H.C.; Neal, D.E. Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene 2009, 28, 3663–3670. [Google Scholar] [CrossRef] [PubMed]
- Ash, S.C.; Yang, D.Q.; Britt, D.E. LYRIC/AEG-1 overexpression modulates BCCIPalpha protein levels in prostate tumor cells. Biochem. Biophys. Res. Commun. 2008, 371, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Thirkettle, H.J.; Girling, J.; Warren, A.Y.; Mills, I.G.; Sahadevan, K.; Leung, H.; Hamdy, F.; Whitaker, H.C.; Neal, D.E. LYRIC/AEG-1 is targeted to different subcellular compartments by ubiquitinylation and intrinsic nuclear localization signals. Clin. Cancer Res. 2009, 15, 3003–3013. [Google Scholar] [CrossRef]
- Packer, J.R.; Maitland, N.J. The molecular and cellular origin of human prostate cancer. Biochim. Biophys. Acta 2016, 1863, 1238–1260. [Google Scholar] [CrossRef]
- Da Costa, P.J.; Menezes, J.; Romao, L. The role of alternative splicing coupled to nonsense-mediated mRNA decay in human disease. Int. J. Biochem. Cell Biol. 2017, 91, 168–175. [Google Scholar] [CrossRef]
- Goncalves, V.; Pereira, J.F.S.; Jordan, P. Signaling Pathways Driving Aberrant Splicing in Cancer Cells. Genes 2017, 9, 9. [Google Scholar] [CrossRef]
- Amin, E.M.; Oltean, S.; Hua, J.; Gammons, M.V.; Hamdollah-Zadeh, M.; Welsh, G.I.; Cheung, M.K.; Ni, L.; Kase, S.; Rennel, E.S.; et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell 2011, 20, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Pelisch, F.; Khauv, D.; Risso, G.; Stallings-Mann, M.; Blaustein, M.; Quadrana, L.; Radisky, D.C.; Srebrow, A. Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J. Cell Biochem. 2012, 113, 2319–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gokmen-Polar, Y.; Murray, N.R.; Velasco, M.A.; Gatalica, Z.; Fields, A.P. Elevated protein kinase C betaII is an early promotive event in colon carcinogenesis. Cancer Res. 2001, 61, 1375–1381. [Google Scholar] [PubMed]
- Nayler, O.; Hartmann, A.M.; Stamm, S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 2000, 150, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; zur Hausen, A.; Orlowska-Volk, M.; Jager, M.; Bettendorf, H.; Stamm, S.; Hirschfeld, M.; Yiqin, O.; Tong, X.; Gitsch, G.; et al. Alternative splicing-related factor YT521: An independent prognostic factor in endometrial cancer. Int. J. Gynecol. Cancer 2010, 20, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Stoilov, P.; Rafalska, I.; Stamm, S. YTH: A new domain in nuclear proteins. Trends Biochem. Sci. 2002, 27, 495–497. [Google Scholar] [CrossRef]
- Hartmann, A.M.; Nayler, O.; Schwaiger, F.W.; Obermeier, A.; Stamm, S. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. Cell 1999, 10, 3909–3926. [Google Scholar] [CrossRef] [PubMed]
- Rafalska, I.; Zhang, Z.; Benderska, N.; Wolff, H.; Hartmann, A.M.; Brack-Werner, R.; Stamm, S. The intranuclear localization and function of YT521-B is regulated by tyrosine phosphorylation. Hum. Mol. Genet. 2004, 13, 1535–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschfeld, M.; Zhang, B.; Jaeger, M.; Stamm, S.; Erbes, T.; Mayer, S.; Tong, X.; Stickeler, E. Hypoxia-dependent mRNA expression pattern of splicing factor YT521 and its impact on oncological important target gene expression. Mol. Carcinog 2014, 53, 883–892. [Google Scholar] [CrossRef]
- Zhang, B.; Shao, X.; Zhou, J.; Qiu, J.; Wu, Y.; Cheng, J. YT521 promotes metastases of endometrial cancer by differential splicing of vascular endothelial growth factor A. Tumor Biol. 2015, 37, 15543–15549. [Google Scholar] [CrossRef]
- Matter, N.; Herrlich, P.; Konig, H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 2002, 420, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Valacca, C.; Bonomi, S.; Buratti, E.; Pedrotti, S.; Baralle, F.E.; Sette, C.; Ghigna, C.; Biamonti, G. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 2010, 191, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Bielli, P.; Busa, R.; Paronetto, M.P.; Sette, C. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr. Relat. Cancer 2011, 18, R91–R102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busa, R.; Geremia, R.; Sette, C. Genotoxic stress causes the accumulation of the splicing regulator Sam68 in nuclear foci of transcriptionally active chromatin. Nucleic Acids Res. 2010, 38, 3005–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Boisvert, F.M.; Bazett-Jones, D.P.; Richard, S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell 1999, 10, 3015–3033. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. Review: Perinucleolar structures. J. Struct. Biol. 2000, 129, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Fong, K.W.; Li, Y.; Wang, W.; Ma, W.; Li, K.; Qi, R.Z.; Liu, D.; Songyang, Z.; Chen, J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J. Cell Biol. 2013, 203, 149–164. [Google Scholar] [CrossRef]
- Britt, D.E.; Yang, D.F.; Yang, D.Q.; Flanagan, D.; Callanan, H.; Lim, Y.P.; Lin, S.H.; Hixson, D.C. Identification of a novel protein, LYRIC, localized to tight junctions of polarized epithelial cells. Exp. Cell Res. 2004, 300, 134–148. [Google Scholar] [CrossRef]
- Rajan, P.; Gaughan, L.; Dalgliesh, C.; El-Sherif, A.; Robson, C.N.; Leung, H.Y.; Elliott, D.J. Regulation of gene expression by the RNA-binding protein Sam68 in cancer. Biochem. Soc. Trans. 2008, 36, 505–507. [Google Scholar] [CrossRef]
- Rajan, P.; Gaughan, L.; Dalgliesh, C.; El-Sherif, A.; Robson, C.N.; Leung, H.Y.; Elliott, D.J. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J. Pathol. 2008, 215, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Ehrmann, I.; Fort, P.; Elliott, D.J. STARs in the CNS. Biochem. Soc. Trans. 2016, 44, 1066–1072. [Google Scholar] [CrossRef] [PubMed]
- Huot, M.E.; Vogel, G.; Zabarauskas, A.; Ngo, C.T.; Coulombe-Huntington, J.; Majewski, J.; Richard, S. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol. Cell 2012, 46, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Vogel, G.; Richard, S. Emerging roles for Sam68 in adipogenesis and neuronal development. RNA Biol. 2012, 9, 1129–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivier, J.; Stavrinides, V.; Kay, J.; Freeman, A.; Pye, H.; Ahmed, Z.; Carmona Echeverria, L.; Heavey, S.; Simmons, L.A.M.; Kanthabalan, A.; et al. Immunohistochemical biomarker validation in highly selective needle biopsy microarrays derived from mpMRI-characterized prostates. Prostate 2018, 78, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Emdad, L.; Sarkar, D.; Su, Z.Z.; Randolph, A.; Boukerche, H.; Valerie, K.; Fisher, P.B. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Res. 2006, 66, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Sernbo, S.; Borrebaeck, C.A.; Uhlen, M.; Jirstrom, K.; Ek, S. Nuclear T-STAR protein expression correlates with HER2 status, hormone receptor negativity and prolonged recurrence free survival in primary breast cancer and decreased cancer cell growth in vitro. PLoS ONE 2013, 8, e70596. [Google Scholar] [CrossRef] [PubMed]
- Richard, S.; Vogel, G.; Huot, M.E.; Guo, T.; Muller, W.J.; Lukong, K.E. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene 2008, 27, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.I. Splicing Factors Have an Essential Role in Prostate Cancer Progression and Androgen Receptor Signaling. Biomolecules 2019, 9, 131. [Google Scholar] [CrossRef]
- Harada, T.; Abe, T.; Kato, F.; Matsumoto, R.; Fujita, H.; Murai, S.; Miyajima, N.; Tsuchiya, K.; Maruyama, S.; Kudo, K.; et al. Five-point Likert scaling on MRI predicts clinically significant prostate carcinoma. BMC Urol. 2015, 15, 91. [Google Scholar] [CrossRef]
- Li, P.; You, S.; Nguyen, C.; Wang, Y.; Kim, J.; Sirohi, D.; Ziembiec, A.; Luthringer, D.; Lin, S.C.; Daskivich, T.; et al. Genes involved in prostate cancer progression determine MRI visibility. Theranostics 2018, 8, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Stoyanova, R.; Pollack, A.; Takhar, M.; Lynne, C.; Parra, N.; Lam, L.L.; Alshalalfa, M.; Buerki, C.; Castillo, R.; Jorda, M.; et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget 2016, 7, 53362–53376. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, G.; Srivastava, N.; Goyal, M.; Rakha, E.; Lothion-Roy, J.; Mongan, N.P.; Miftakhova, R.R.; Khaiboullina, S.F.; Rizvanov, A.A.; Baranwal, M. Metadherin: A Therapeutic Target in Multiple Cancers. Front. Oncol. 2019, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, H.G.; Lam, Y.W.; Briers, S.; Lamond, A.I.; Bickmore, W.A. 3D3/lyric: A novel transmembrane protein of the endoplasmic reticulum and nuclear envelope, which is also present in the nucleolus. Exp. Cell Res. 2004, 294, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Weg-Remers, S.; Ponta, H.; Herrlich, P.; Konig, H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001, 20, 4194–4203. [Google Scholar] [CrossRef] [PubMed]
- Ishidate, T.; Yoshihara, S.; Kawasaki, Y.; Roy, B.C.; Toyoshima, K.; Akiyama, T. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 1997, 409, 237–241. [Google Scholar] [CrossRef]
- Mizushima, S.; Nagata, S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990, 18, 5322. [Google Scholar] [CrossRef]
- Whitaker, H.C.; Stanbury, D.P.; Brinham, C.; Girling, J.; Hanrahan, S.; Totty, N.; Neal, D.E. Labeling and identification of LNCaP cell surface proteins: A pilot study. Prostate 2007, 67, 943–954. [Google Scholar] [CrossRef]
- Fedchenko, N.; Reifenrath, J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue—A review. Diagn. Pathol. 2014, 9, 221. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luxton, H.J.; Simpson, B.S.; Mills, I.G.; Brindle, N.R.; Ahmed, Z.; Stavrinides, V.; Heavey, S.; Stamm, S.; Whitaker, H.C. The Oncogene Metadherin Interacts with the Known Splicing Proteins YTHDC1, Sam68 and T-STAR and Plays a Novel Role in Alternative mRNA Splicing. Cancers 2019, 11, 1233. https://doi.org/10.3390/cancers11091233
Luxton HJ, Simpson BS, Mills IG, Brindle NR, Ahmed Z, Stavrinides V, Heavey S, Stamm S, Whitaker HC. The Oncogene Metadherin Interacts with the Known Splicing Proteins YTHDC1, Sam68 and T-STAR and Plays a Novel Role in Alternative mRNA Splicing. Cancers. 2019; 11(9):1233. https://doi.org/10.3390/cancers11091233
Chicago/Turabian StyleLuxton, Hayley J., Benjamin S. Simpson, Ian G. Mills, Nicola R. Brindle, Zeba Ahmed, Vasilis Stavrinides, Susan Heavey, Stefan Stamm, and Hayley C. Whitaker. 2019. "The Oncogene Metadherin Interacts with the Known Splicing Proteins YTHDC1, Sam68 and T-STAR and Plays a Novel Role in Alternative mRNA Splicing" Cancers 11, no. 9: 1233. https://doi.org/10.3390/cancers11091233