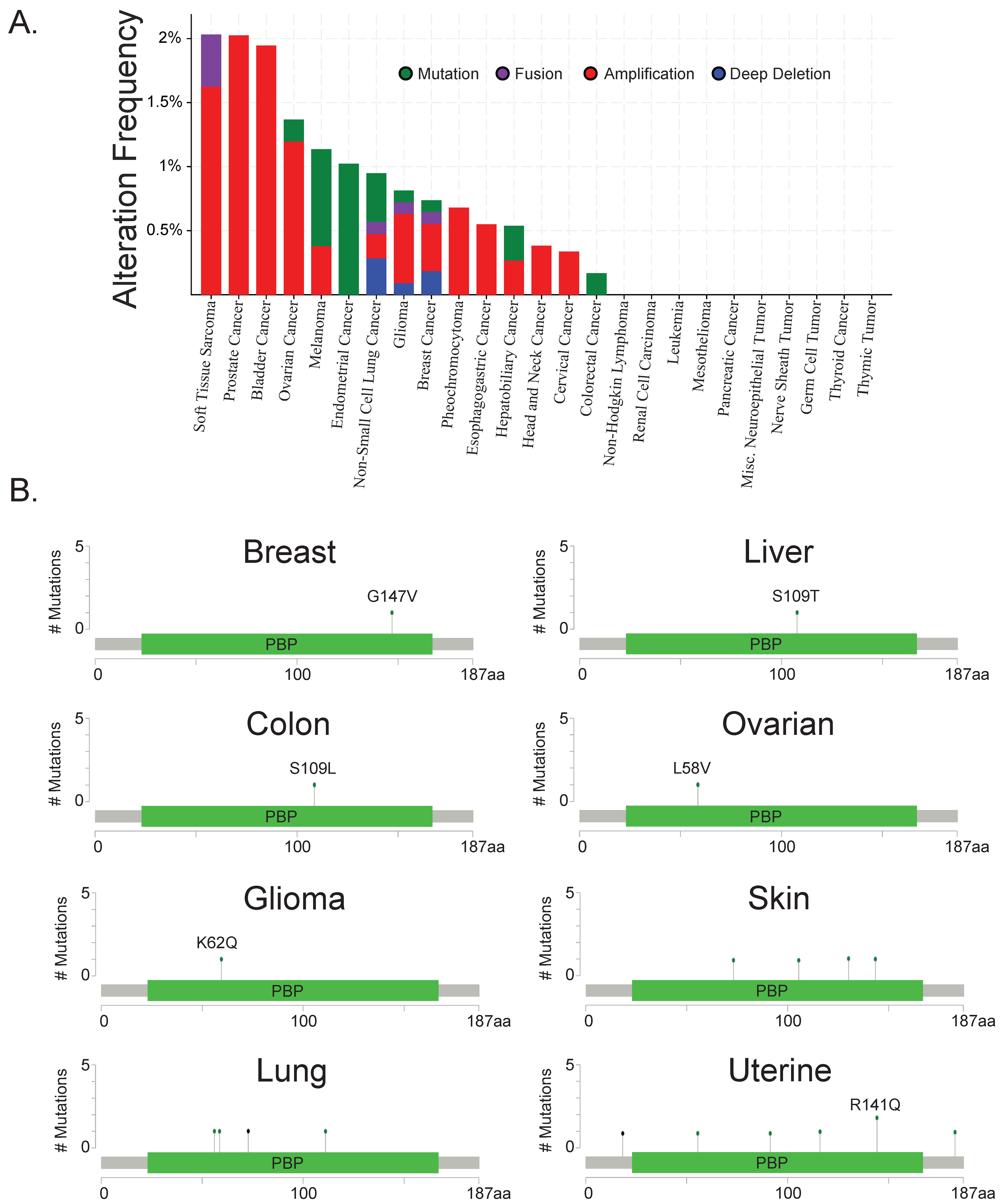
Targeting Raf Kinase Inhibitory Protein Regulation and Function
Abstract
:1. Introduction
2. Mechanisms of RKIP Downregulation and Strategies for Recovering RKIP Expression
2.1. Epigenetic Silencing Through Promoter Methylation and Histone Modifications
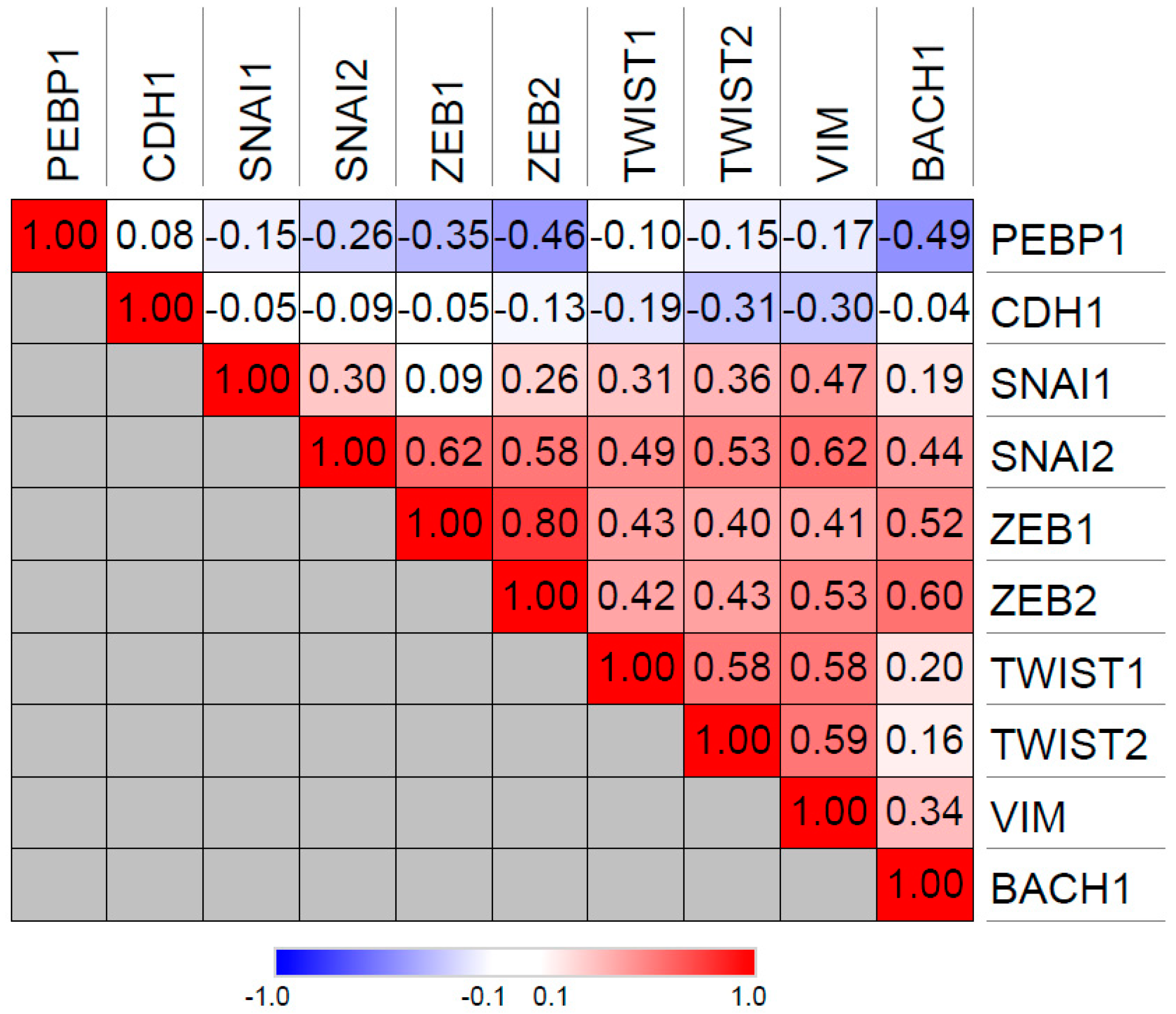
2.2. Transcriptional Regulation of RKIP
2.3. Post-Transcriptional Regulation of RKIP
2.4. Chemical Induction of RKIP
2.5. Regulation and Function of RKIP in Liquid Cancers
3. Identifying Downstream Networks to Mimic RKIP Function
3.1. Kinase Targets
3.2. Mimicking Microenvironmental Effects of RKIP in Tumors
3.3. Computational Approaches to Build Actionable RKIP Gene Networks
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Lamiman, K.; Keller, J.M.; Mizokami, A.; Zhang, J.; Keller, E.T. Survey of Raf kinase inhibitor protein (RKIP) in multiple cancer types. Crit. Rev. Oncog. 2014, 19, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Martinho, O.; Granja, S.; Jaraquemada, T.; Caeiro, C.; Miranda-Gonçalves, V.; Honavar, M.; Costa, P.; Damasceno, M.; Rosner, M.R.; Lopes, J.M.; et al. Downregulation of RKIP is associated with poor outcome and malignant progression in gliomas. PLoS ONE 2012, 7, e30769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinho, O.; Pinto, F.; Granja, S.; Miranda-Gonçalves, V.; Moreira, M.A.R.; Ribeiro, L.F.J.; di Loreto, C.; Rosner, M.R.; Longatto-Filho, A.; Reis, R.M. RKIP inhibition in cervical cancer is associated with higher tumor aggressive behavior and resistance to cisplatin therapy. PLoS ONE 2013, 8, e59104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangi-Garimella, S.; Yun, J.; Eves, E.M.; Newman, M.; Erkeland, S.J.; Hammond, S.M.; Minn, A.J.; Rosner, M.R. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.; Smith, P.C.; Zhang, L.; Rubin, M.A.; Dunn, R.L.; Yao, Z.; Keller, E.T. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J. Natl. Cancer Inst. 2003, 95, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Bainer, R.; Frankenberger, C.; Rabe, D.; An, G.; Gilad, Y.; Rosner, M.R. Gene expression in local stroma reflects breast tumor states and predicts patient outcome. Sci. Rep. 2016, 6, e39240. [Google Scholar] [CrossRef] [PubMed]
- Frankenberger, C.; Rabe, D.; Bainer, R.; Sankarasharma, D.; Chada, K.; Krausz, T.; Gilad, Y.; Becker, L.; Rosner, M.R. Metastasis Suppressors Regulate the Tumor Microenvironment by Blocking Recruitment of Prometastatic Tumor-Associated Macrophages. Cancer Res. 2015, 75, 4063–4073. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Frankenberger, C.; Yun, J.; Bevilacqua, E.; Caldas, C.; Chin, S.-F.; Rueda, O.M.; Reinitz, J.; Rosner, M.R. A prognostic gene signature for metastasis-free survival of triple negative breast cancer patients. PLoS ONE 2013, 8, e82125. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Song, C.-X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, J.; Frankenberger, C.A.; Kuo, W.-L.; Boelens, M.C.; Eves, E.M.; Cheng, N.; Liang, H.; Li, W.-H.; Ishwaran, H.; Minn, A.J.; et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011, 30, 4500–4514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Yesilkanal, A.E.; Rosner, M.R. Raf kinase inhibitory protein (RKIP) as a metastasis suppressor: Regulation of signaling networks in cancer. Crit. Rev. Oncog. 2014, 19, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wenzel, S. Interactions of RKIP with inflammatory signaling pathways. Crit. Rev. Oncog. 2014, 19, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.; Lohse, M.J.; Quitterer, U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 2003, 426, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.J.; Rosner, M.R. RKIP Structure Drives Its Function: A Three-State Model for Regulation of RKIP. Crit. Rev. Oncog. 2014, 19, 483–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinner, J.J.; Wang, S.; Lee, J.; Ong, C.; Sommese, R.; Sivaramakrishnan, S.; Koelmel, W.; Hirschbeck, M.; Schindelin, H.; Kisker, C.; et al. Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome. Proc. Natl. Acad. Sci. USA. 2017, 114, 13453–13458. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Dong, Z.; Guo, Y.; Lin, X.; Chen, Z.; Kuang, G.; Yang, Z. Aberrant methylation and loss expression of RKIP is associated with tumor progression and poor prognosis in gastric cardia adenocarcinoma. Clin. Exp. Metastasis 2013, 30, 265–275. [Google Scholar] [CrossRef] [PubMed]
- LI, D.-X.; CAI, H.-Y.; WANG, X.; FENG, Y.-L.; CAI, S.-W. Promoter methylation of Raf kinase inhibitory protein: A significant prognostic indicator for patients with gastric adenocarcinoma. Exp. Ther. Med. 2014, 8, 844–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Dong, Z.; Lin, X.; Zhang, M.; Kuang, G.; Zhu, T. Decreased expression and aberrant methylation of Raf kinase inhibitory protein gene in esophageal squamous cell carcinoma. Cancer Investig. 2012, 30, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Liu, Z.; She, H.; Liu, B.; Gu, J.; Wei, D.; Zhang, X.; Wang, J.; Qi, S.; Ping, F. Promoter methylation and expression of Raf kinase inhibitory protein in esophageal squamous cell carcinoma. Oncol. Lett. 2017, 13, 1866–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Mulla, F.; Hagan, S.; Al-Ali, W.; Jacob, S.P.; Behbehani, A.I.; Bitar, M.S.; Dallol, A.; Kolch, W. Raf kinase inhibitor protein: mechanism of loss of expression and association with genomic instability. J. Clin. Pathol. 2008, 61, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Minoo, P.; Baker, K.; Goswami, R.; Chong, G.; Foulkes, W.D.; Ruszkiewicz, A.R.; Barker, M.; Buchanan, D.; Young, J.; Jass, J.R. Extensive DNA methylation in normal colorectal mucosa in hyperplastic polyposis. Gut. 2006, 55, 1467–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.-E.; Kim, N.I.; Lee, J.S.; Park, M.H.; Yoon, J.H. Reduced RKIP Expression is Associated with Breast Neoplastic Progression and is Correlated with Poor Outcomes and Aberrant Methylation in Breast Carcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM 2017, 25, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Beach, S.; Tang, H.; Park, S.; Dhillon, A.S.; Keller, E.T.; Kolch, W.; Yeung, K.C. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene 2008, 27, 2243–2248. [Google Scholar] [CrossRef] [PubMed]
- Labbozzetta, M.; Poma, P.; Vivona, N.; Gulino, A.; D’Alessandro, N.; Notarbartolo, M. Epigenetic changes and nuclear factor-κB activation, but not microRNA-224, downregulate Raf-1 kinase inhibitor protein in triple-negative breast cancer SUM 159 cells. Oncol. Lett. 2015, 10, 3807–3815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Lee, J.; Farquhar, K.S.; Yun, J.; Frankenberger, C.A.; Bevilacqua, E.; Yeung, K.; Kim, E.-J.; Balázsi, G.; Rosner, M.R. Network of mutually repressive metastasis regulators can promote cell heterogeneity and metastatic transitions. Proc. Natl. Acad. Sci. USA 2014, 111, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Baritaki, S.; Marathe, H.; Feng, J.; Park, S.; Beach, S.; Bazeley, P.S.; Beshir, A.B.; Fenteany, G.; Mehra, R.; et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012, 72, 3091–3104. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B.; Baritaki, S. Dual role of NO donors in the reversal of tumor cell resistance and EMT: Downregulation of the NF-κB/Snail/YY1/RKIP circuitry. Nitric Oxide Biol. Chem. 2011, 24, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Bhutia, S.K.; Sokhi, U.K.; Azab, B.; Su, Z.; Boukerche, H.; Anwar, T.; Moen, E.L.; Chatterjee, D.; Pellecchia, M.; et al. Raf kinase inhibitor RKIP inhibits MDA-9/syntenin-mediated metastasis in melanoma. Cancer Res. 2012, 72, 6217–6226. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wu, H.; Lei, R.; Chong, R.A.; Wei, Y.; Lu, X.; Tagkopoulos, I.; Kung, S.-Y.; Yang, Q.; Hu, G.; et al. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J. Biol. Chem. 2012, 287, 33533–33544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, J.; Keller, J.M.; Yeung, K.; Keller, E.T.; Fu, Z. Transcriptional regulation of RKIP expression by androgen in prostate cells. Cell Physiol. Biochem. Int. J. Exp. Cell Physiol. Biochem. Pharmacol. 2012, 30, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, O.; Qin, J.; Liu, S.; Sun, S.; Liu, H.; Kuang, J.; Jiang, G.; Zhang, W. cis-Acting elements and trans-acting factors in the transcriptional regulation of raf kinase inhibitory protein expression. PLoS ONE 2013, 8, e83097. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Song, Y.; Fu, Z.; Yu, W. miR-27a regulates cisplatin resistance and metastasis by targeting RKIP in human lung adenocarcinoma cells. Mol. Cancer 2014, 13, e193. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, X.-H.; Zhu, H.-C.; Wang, L.; Ning, J.-Z.; Xiao, C.-C. MiR-543 Promotes Proliferation and Epithelial-Mesenchymal Transition in Prostate Cancer via Targeting RKIP. Cell Physiol. Biochem. Int. J. Exp. Cell Physiol. Biochem. Pharmacol. 2017, 41, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Weng, X.-D.; Wang, L.; Liu, X.-H.; Zhu, H.-C.; Guo, J.; Ning, J.-Z.; Xiao, C.-C. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget 2017, 8, 94358–94370. [Google Scholar] [CrossRef] [PubMed]
- Hatzl, S.; Geiger, O.; Kuepper, M.K.; Caraffini, V.; Seime, T.; Furlan, T.; Nussbaumer, E.; Wieser, R.; Pichler, M.; Scheideler, M.; et al. Increased Expression of miR-23a Mediates a Loss of Expression in the RAF Kinase Inhibitor Protein RKIP. Cancer Res. 2016, 76, 3644–3654. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Dai, T.; Lin, X.; Zhao, X.; Chen, X.; Wang, C.; Li, X.; Shen, H.; Wang, X. MicroRNA-224 targets RKIP to control cell invasion and expression of metastasis genes in human breast cancer cells. Biochem. Biophys. Res. Commun. 2012, 425, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Poma, P.; Labbozzetta, M.; Vivona, N.; Porcasi, R.; D’Alessandro, N.; Notarbartolo, M. Analysis of possible mechanisms accounting for raf-1 kinase inhibitor protein downregulation in hepatocellular carcinoma. Omics J. Integr. Biol. 2012, 16, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Choi, G.H.; Jung, Y.; Kim, K.M.; Jang, S.-J.; Yu, E.S.; Lee, H.C. Downregulation of Raf-1 kinase inhibitory protein as a sorafenib resistance mechanism in hepatocellular carcinoma cell lines. J. Cancer Res. Clin. Oncol. 2018, 144, 1487–1501. [Google Scholar] [CrossRef] [PubMed]
- Moen, E.L.; Wen, S.; Anwar, T.; Cross-Knorr, S.; Brilliant, K.; Birnbaum, F.; Rahaman, S.; Sedivy, J.M.; Moss, S.F.; Chatterjee, D. Regulation of RKIP function by Helicobacter pylori in gastric cancer. PLoS ONE 2012, 7, e37819. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Shu, Y.; Gao, C.; Wang, X.; Qi, G.; Zhang, P.; Li, M.; Shi, J.; Tian, B. CDK5-mediated phosphorylation and autophagy of RKIP regulate neuronal death in Parkinson’s disease. Neurobiol. Aging 2014, 35, 2870–2880. [Google Scholar] [CrossRef] [PubMed]
- Baritaki, S.; Huerta-Yepez, S.; Sahakyan, A.; Karagiannides, I.; Bakirtzi, K.; Jazirehi, A.; Bonavida, B. Mechanisms of nitric oxide-mediated inhibition of EMT in cancer: Inhibition of the metastasis-inducer Snail and induction of the metastasis-suppressor RKIP. Cell Cycle 2010, 9, 4931–4940. [Google Scholar] [CrossRef] [PubMed]
- Tsao, D.-A.; Yu, H.-S.; Chang, H.-R. Nitric oxide enhances expression of raf kinase inhibitor protein in keratinocytes. Exp. Dermatol. 2009, 18, 571–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualetti, G.; Ricciardi, S.; Mey, V.; Del Tacca, M.; Danesi, R. Synergistic cytotoxicity, inhibition of signal transduction pathways and pharmacogenetics of sorafenib and gemcitabine in human NSCLC cell lines. Lung Cancer Amst. Neth. 2011, 74, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, S.; Mey, V.; Nannizzi, S.; Pasqualetti, G.; Crea, F.; Del Tacca, M.; Danesi, R. Synergistic cytotoxicity and molecular interaction on drug targets of sorafenib and gemcitabine in human pancreas cancer cells. Chemotherapy 2010, 56, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Labots, M.; Dekker, H.; Galvani, E.; Lind, J.S.W.; Sciarrillo, R.; Honeywell, R.; Smit, E.F.; Verheul, H.M.; Peters, G.J. Molecular mechanisms and modulation of key pathways underlying the synergistic interaction of sorafenib with erlotinib in non-small-cell-lung cancer (NSCLC) cells. Curr. Pharm. Des. 2013, 19, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Bai, Y.; Wang, Z.; Beach, S.; Mott, S.; Roy, R.; Braastad, C.; Sun, Y.; Mukhopadhyay, A.; Aggarwal, B.B.; et al. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J. Biol. Chem. 2004, 279, 17515–17523. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, S.; Duan, M.; Moen, E.L.; Cross-Knorr, S.; Brilliant, K.; Bonavida, B.; LaValle, T.; Yeung, K.C.; Al-Mulla, F.; Chin, E.; et al. Raf kinase inhibitor protein (RKIP) blocks signal transducer and activator of transcription 3 (STAT3) activation in breast and prostate cancer. PLoS ONE 2014, 9, e92478. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-C.; Hsu, Y.-L.; Tsai, Y.-C.; Chang, Y.-W.; Kuo, P.-L.; Chen, Y.-H. Gemifloxacin inhibits migration and invasion and induces mesenchymal-epithelial transition in human breast adenocarcinoma cells. J. Mol. Med. Berl. Ger. 2014, 92, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Beshir, A.B.; Argueta, C.E.; Menikarachchi, L.C.; Gascón, J.A.; Fenteany, G. Locostatin Disrupts Association of Raf Kinase Inhibitor Protein with Binding Proteins by Modifying a Conserved Histidine Residue in the Ligand-Binding Pocket. Forum Immunopathol. Dis. Ther. 2011, 2, 47–58. [Google Scholar] [CrossRef]
- Zhu, S.; Mc Henry, K.T.; Lane, W.S.; Fenteany, G. A chemical inhibitor reveals the role of Raf kinase inhibitor protein in cell migration. Chem. Biol. 2005, 12, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Shemon, A.N.; Eves, E.M.; Clark, M.C.; Heil, G.; Granovsky, A.; Zeng, L.; Imamoto, A.; Koide, S.; Rosner, M.R. Raf Kinase Inhibitory Protein protects cells against locostatin-mediated inhibition of migration. PLoS ONE 2009, 4, e6028. [Google Scholar] [CrossRef] [PubMed]
- Zebisch, A.; Wölfler, A.; Fried, I.; Wolf, O.; Lind, K.; Bodner, C.; Haller, M.; Drasche, A.; Pirkebner, D.; Matallanas, D.; et al. Frequent loss of RAF kinase inhibitor protein expression in acute myeloid leukemia. Leukemia 2012, 26, 1842–1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caraffini, V.; Perfler, B.; Berg, J.L.; Uhl, B.; Schauer, S.; Kashofer, K.; Ghaffari-Tabrizi-Wizsy, N.; Strobl, H.; Wölfler, A.; Hoefler, G.; et al. Loss of RKIP is a frequent event in myeloid sarcoma and promotes leukemic tissue infiltration. Blood 2018, 131, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Stahl, M.; Kohrman, N.; Gore, S.D.; Kim, T.K.; Zeidan, A.M.; Prebet, T. Epigenetics in Cancer: A Hematological Perspective. PLoS Genet. 2016, 12, e1006193. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jiang, X.; Li, Z.; Li, Y.; Song, C.-X.; He, C.; Sun, M.; Chen, P.; Gurbuxani, S.; Wang, J.; et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc. Natl. Acad. Sci. USA. 2013, 110, 11994–11999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baritaki, S.; Huerta-Yepez, S.; da Lourdas Cabrava-Haimandez, M.; Sensi, M.; Canevari, S.; Libra, M.; Penichet, M.; Chen, H.; Berenson, J.R.; Bonavida, B. Unique Pattern of Overexpression of Raf-1 Kinase Inhibitory Protein in Its Inactivated Phosphorylated Form in Human Multiple Myeloma. Forum Immunopathol. Dis. Ther. 2011, 2, 179–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jazirehi, A.R.; Vega, M.I.; Chatterjee, D.; Goodglick, L.; Bonavida, B. Inhibition of the Raf-MEK1/2-ERK1/2 signaling pathway, Bcl-xL down-regulation, and chemosensitization of non-Hodgkin’s lymphoma B cells by Rituximab. Cancer Res. 2004, 64, 7117–7126. [Google Scholar] [CrossRef] [PubMed]
- Vega, M.I.; Martínez-Paniagua, M.; Huerta-Yepez, S.; González-Bonilla, C.; Uematsu, N.; Bonavida, B. Dysregulation of the cell survival/anti-apoptotic NF-κB pathway by the novel humanized BM-ca anti-CD20 mAb: Implication in chemosensitization. Int. J. Oncol. 2009, 35, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Baritaki, S.; Militello, L.; Malaponte, G.; Spandidos, D.A.; Salcedo, M.; Bonavida, B. The anti-CD20 mAb LFB-R603 interrupts the dysregulated NF-κB/Snail/RKIP/PTEN resistance loop in B-NHL cells: Role in sensitization to TRAIL apoptosis. Int. J. Oncol. 2011, 38, 1683–1694. [Google Scholar] [PubMed]
- Trakul, N.; Menard, R.E.; Schade, G.R.; Qian, Z.; Rosner, M.R. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J. Biol. Chem. 2005, 280, 24931–24940. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.; Janosch, P.; McFerran, B.; Rose, D.W.; Mischak, H.; Sedivy, J.M.; Kolch, W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the raf kinase inhibitor protein. Mol. Cell. Biol. 2000, 20, 3079–3085. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.C.; Rose, D.W.; Dhillon, A.S.; Yaros, D.; Gustafsson, M.; Chatterjee, D.; McFerran, B.; Wyche, J.; Kolch, W.; Sedivy, J.M. Raf kinase inhibitor protein interacts with NF-κB-inducing kinase and TAK1 and inhibits NF-κB activation. Mol. Cell. Biol. 2001, 21, 7207–7217. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.M.; Simeone, E.; Festino, L.; Vanella, V.; Strudel, M.; Ascierto, P.A. MEK Inhibitors in the Treatment of Metastatic Melanoma and Solid Tumors. Am. J. Clin. Dermatol. 2017, 18, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Battula, V.L.; Nguyen, K.; Sun, J.; Pitner, M.K.; Yuan, B.; Bartholomeusz, C.; Hail, N.; Andreeff, M. IKK inhibition by BMS-345541 suppresses breast tumorigenesis and metastases by targeting GD2+ cancer stem cells. Oncotarget 2017, 8, 36936–36949. [Google Scholar] [CrossRef] [PubMed]
- Andela, V.B.; Schwarz, E.M.; Puzas, J.E.; O’Keefe, R.J.; Rosier, R.N. Tumor Metastasis and the Reciprocal Regulation of Prometastatic and Antimetastatic Factors by Nuclear Factor κB. Cancer Res. 2000, 60, 6557–6562. [Google Scholar] [PubMed]
- Golubovskaya, V.M. Targeting FAK in human cancer: From finding to first clinical trials. Front. Biosci. Landmark Ed. 2014, 19, 687–706. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.; Logothetis, C. Dasatinib: A potent SRC inhibitor in clinical development for the treatment of solid tumors. Cancer Treat. Rev. 2010, 36, 492–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, J.S.; Whittle, M.C.; Nakamura, K.; Abell, A.N.; Midland, A.A.; Zawistowski, J.S.; Johnson, N.L.; Granger, D.A.; Jordan, N.V.; Darr, D.B.; et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 2012, 149, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Ideker, T.; Dutkowski, J.; Hood, L. Boosting signal-to-noise in complex biology: Prior knowledge is power. Cell 2011, 144, 860–863. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yesilkanal, A.E.; Rosner, M.R. Targeting Raf Kinase Inhibitory Protein Regulation and Function. Cancers 2018, 10, 306. https://doi.org/10.3390/cancers10090306
Yesilkanal AE, Rosner MR. Targeting Raf Kinase Inhibitory Protein Regulation and Function. Cancers. 2018; 10(9):306. https://doi.org/10.3390/cancers10090306
Chicago/Turabian StyleYesilkanal, Ali Ekrem, and Marsha Rich Rosner. 2018. "Targeting Raf Kinase Inhibitory Protein Regulation and Function" Cancers 10, no. 9: 306. https://doi.org/10.3390/cancers10090306



