Biomass Quantification of the Critically Endangered European eel from Running Waters Using Environmental DNA
Abstract
:1. Introduction
2. Material and Methods
2.1. Real-Time PCR Marker Design and qPCR Procedures
2.1.1. Primer Design
2.1.2. qPCR
2.2. Validation of the qPCR Marker
2.2.1. Experimental Validation in Aquaculture Tanks
2.2.2. Validation in Natural Rivers
- 1.
- Correction for river hydrography
- 2.
- Correlational validation
- 3.
- Predictive value of the new marker
2.2.3. Sampling Procedures in the Field
2.3. eDNA Analysis Procedures
2.3.1. Water Samples Filtration
2.3.2. eDNA Purification
2.3.3. Inhibition Testing and Detection Probability
2.4. End-Time PCR Markers Assayed on Field Samples
2.4.1. Universal Marker
2.4.2. End-Time PCR Eel-Specific Marker
2.5. Data Analysis and Statistics
- -
- Stream order: The number defining the level of river branching at the considered sampling point. It is a discontinuous quantitative variable.
- -
- Altitude: Meters above sea level at each sampling point. It is a continuous quantitative variable.
- -
- Total eDNA quantity: The amount of eDNA in a sample, measured using HS Qubit Fluorometer. It is a continuous quantitative variable.
- -
- End-time PCR eel-specific marker: Using the primers described above from Burgoa Cardás et al. [21], it measures presence/lack of presence of eels’ eDNA (positive amplification/no amplification); thus, the primary variable is binary (0/1). It may be secondarily transformed into a discontinuous quantitative variable using the number of sampling replicates with positive end-time PCR amplification per sampling site (variation range 0–3).
- -
- End-time PCR universal marker: As in the previous case but using Leray et al. [37] universal primers instead of an eel-specific marker.
- -
- Eel eDNA quantity: CT values obtained when amplifying eel eDNA using the species-specific primers developed in the current study. It is a continuous quantitative variable.
- -
- Adjusted eel eDNA quantity: Amount of eel eDNA estimated from field samples as an extrapolation from the standard curve. It is a continuous quantitative variable. In the analysis of field results, CT values were preferred as a proxy to eDNA quantity estimations for being a fitter measurement for regression analysis.
3. Results
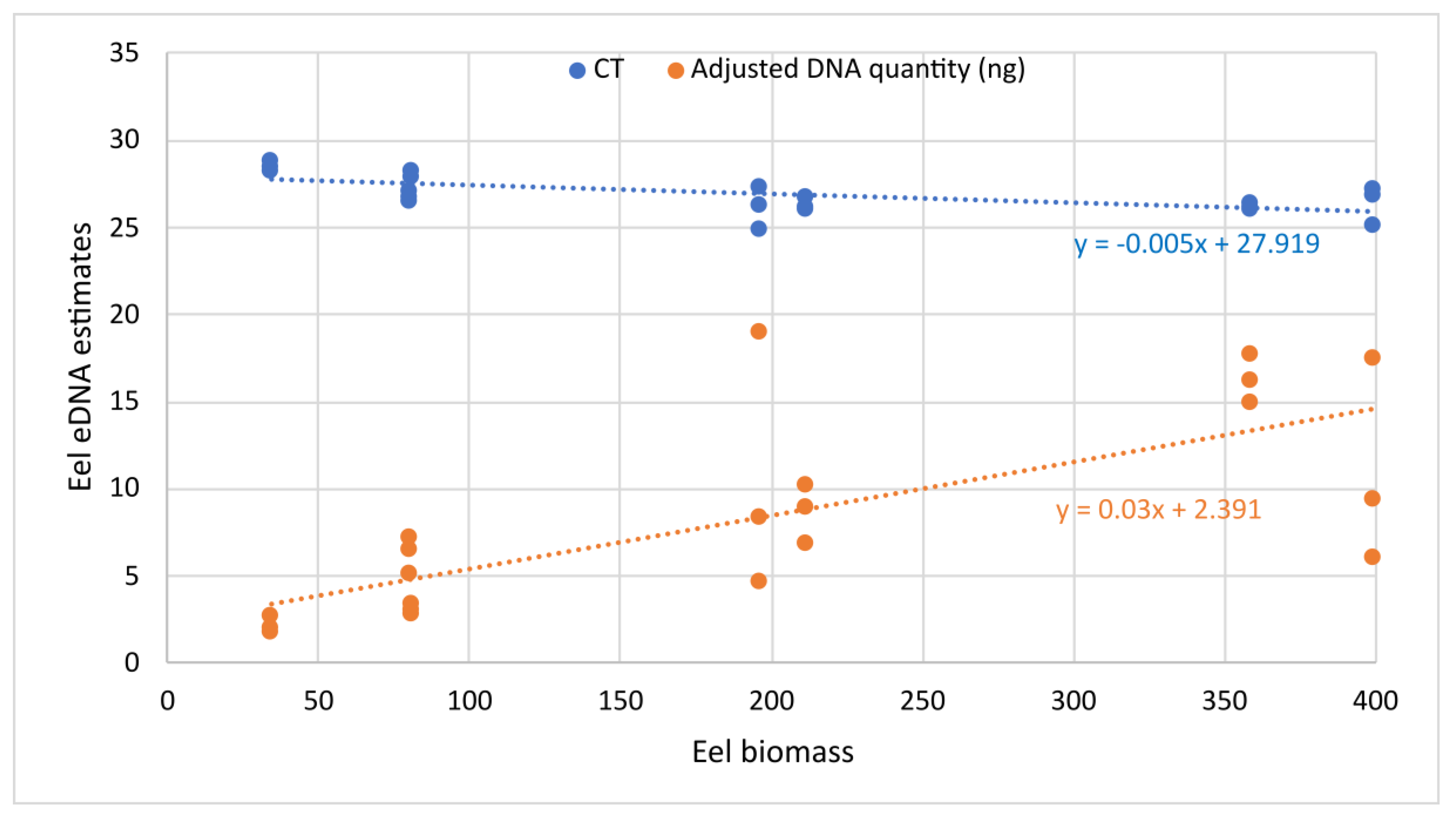
3.1. Primers Validation In Vitro and in Experimental Tanks
| Tank Number | Biomass | Number of Individuals | Tank Sampling Replicate | CT Mean | Eels’ DNA Quantity Per Tank (ng) |
|---|---|---|---|---|---|
| T1 | 34.5 | 3 | A | 28.27 | 0.275 |
| B | 28.81 | 0.176 | |||
| C | 28.49 | 0.197 | |||
| T2 | 80.57 | 4 | A | 27.04 | 0.508 |
| B | 26.52 | 0.723 | |||
| C | 26.70 | 0.651 | |||
| T3 | 81.15 | 4 | A | 28.23 | 0.344 |
| B | 28.29 | 0.286 | |||
| C | 27.91 | 0.310 | |||
| T4 | 196.3 | 8 | A | 26.31 | 0.832 |
| B | 27.31 | 0.465 | |||
| C | 24.93 | 1.902 | |||
| T5 | 211.7 | 14 | A | 26.18 | 0.896 |
| B | 26.74 | 0.691 | |||
| C | 26.04 | 1.027 | |||
| T6 | 358.6 | 20 | A | 26.17 | 1.619 |
| B | 26.10 | 1.772 | |||
| C | 26.44 | 1.496 | |||
| T7 | 399.5 | 24 | A | 25.13 | 1.750 |
| B | 26.86 | 0.608 | |||
| C | 27.15 | 0.945 | |||
| T8 | 745.5 | 43 | A | 28.60 | 0.459 |
| B | 28.68 | 0.392 | |||
| C | 27.41 | 0.857 | |||
| T9 | 748.6 | 43 | A | 27.12 | 0.935 |
| B | 27.58 | 0.778 | |||
| C | 28.19 | 0.490 |
3.2. Field Validation
3.2.1. eDNA Detection and River Hydrography
3.2.2. Relationship between Eel Biomass and Eel eDNA

3.2.3. Predictive Value of the New Marker
4. Discussion
4.1. Overview of the Innovations of the New qPCR Marker
4.2. Limitations of This Study
5. Management Recommendations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feunteun, E. Management and Restoration of European Eel Population (Anguilla anguilla): An Impossible Bargain. Ecol. Eng. 2002, 18, 575–591. [Google Scholar] [CrossRef]
- Bevacqua, D.; Melià, P.; Di Milano, P. Assessing Management Plans for the Recovery of the European Eel: A Need for Multi-Objective Analyses. Am. Fish. Soc. Symp. 2009, 69, 637–647. [Google Scholar]
- Székely, C.; Palstra, A.; Molnár, K.; van den Thillart, G. Impact of the Swim-Bladder Parasite on the Health and Performance of European Eels. In Spawning Migration of the European Eel; Springer: Berlin/Heidelberg, Germany, 2009; pp. 201–226. [Google Scholar] [CrossRef]
- Jacoby, D.; Gollock, M. Anguilla anguilla. The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2014. [Google Scholar]
- Clavero, M.; Hermoso, V. Historical Data to Plan the Recovery of the European Eel. J. Appl. Ecol. 2015, 52, 960–968. [Google Scholar] [CrossRef]
- Aarestrup, K.; Økland, F.; Hansen, M.M.; Righton, D.; Gargan, P.; Castonguay, M.; Bernatchez, L.; Howey, P.; Sparholt, H.; Pedersen, M.I.; et al. Oceanic Spawning Migration of the European Eel (Anguilla anguilla). Science 2009, 325, 1660. [Google Scholar] [CrossRef] [PubMed]
- Dekker, W. The History of Commercial Fisheries for European Eel Commenced Only a Century Ago. Fish. Manag. Ecol. 2019, 26, 6–19. [Google Scholar] [CrossRef]
- Dekker, W. A Conceptual Management Framework for the Restoration of the Declining European Eel Stock. Am. Fish. Soc. Symp. 2009, 58, 3–19. [Google Scholar]
- European Union. Establishing Measures for the Recovery of the Stock of European eel in E Union. 1100/2007; European Union: Maastricht, The Netherlands, 2007. [Google Scholar]
- European Union. Official Journal of the European Union; European Union: Maastricht, The Netherlands, 2007; Volume 248, pp. 281–304. [Google Scholar]
- Weldon, L.; O’Leary, C.; Steer, M.; Newton, L.; Macdonald, H.; Sargeant, S.L. A Comparison of European Eel Anguilla anguilla eDNA Concentrations to Fyke Net Catches in Five Irish Lakes. Environ. DNA 2020, 2, 587–600. [Google Scholar] [CrossRef]
- Lobón-Cerviá, J.; Iglesias, T. Long-Term Numerical Changes and Regulation in a River Stock of European Eel Anguilla anguilla. Freshw. Biol. 2008, 53, 1832–1844. [Google Scholar] [CrossRef]
- Bilotta, G.S.; Sibley, P.; Hateley, J.; Don, A. The Decline of the European Eel Anguilla anguilla: Quantifying and Managing Escapement to Support Conservation. J. Fish Biol. 2011, 78, 23–38. [Google Scholar] [CrossRef]
- Weltersbach, M.S.; Strehlow, H.V.; Ferter, K.; Klefoth, T.; de Graaf, M.; Dorow, M. Estimating and Mitigating Post-Release Mortality of European Eel by Combining Citizen Science with a Catch-and-Release Angling Experiment. Fish. Res. 2018, 201, 98–108. [Google Scholar] [CrossRef]
- Nielsen, J.L. Scientific Sampling Effects: Electrofishing California’s Endangered Fish Populations. Fisheries 1998, 23, 6–12. [Google Scholar] [CrossRef]
- Ellender, B.R.; Becker, A.; Weyl, O.L.F.; Swartz, E.R. Underwater Video Analysis as a Non-Destructive Alternative to Electrofishing for Sampling Imperilled Headwater Stream Fishes. Aquat. Conserv. 2012, 22, 58–65. [Google Scholar] [CrossRef]
- Kanno, Y.; Vokoun, J.C.; Dauwalter, D.C.; Hughes, R.M.; Herlihy, A.T.; Maret, T.R.; Patton, T.M. Influence of Rare Species on Electrofishing Distance When Estimating Species Richness of Stream and River Reaches. Trans. Am. Fish. Soc. 2011, 138, 1240–1251. [Google Scholar] [CrossRef]
- Degerman, E.; Tamario, C.; Watz, J.; Nilsson, P.A.; Calles, O. Occurrence and Habitat Use of European Eel (Anguilla anguilla) in Running Waters: Lessons for Improved Monitoring, Habitat Restoration and Stocking. Aquat. Ecol. 2019, 53, 639–650. [Google Scholar] [CrossRef]
- Fernandez, S.; Sandin, M.M.; Beaulieu, P.G.; Clusa, L.; Martinez, J.L.; Ardura, A.; García-Vázquez, E. Environmental DNA for Freshwater Fish Monitoring: Insights for Conservation within a Protected Area. PeerJ 2018, 2018, e4486. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An Emerging Tool in Conservation for Monitoring Past and Present Biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-Unseen” Detection of Rare Aquatic Species Using Environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Burgoa Cardás, J.; Deconinck, D.; Márquez, I.; Peón Torre, P.; Garcia-Vazquez, E.; Machado-Schiaffino, G. New eDNA Based Tool Applied to the Specific Detection and Monitoring of the Endangered European Eel. Biol. Conserv. 2020, 250, 108750. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Côté, G.; Leclerc, V.; Bernatchez, L. Quantifying Relative Fish Abundance with EDNA: A Promising Tool for Fisheries Management. J. Appl. Ecol. 2016, 53, 1148–1157. [Google Scholar] [CrossRef]
- Minamoto, T.; Miya, M.; Sado, T.; Seino, S.; Doi, H.; Kondoh, M.; Nakamura, K.; Takahara, T.; Yamamoto, S.; Yamanaka, H.; et al. An Illustrated Manual for Environmental DNA Research: Water Sampling Guidelines and Experimental Protocols. Environ. DNA 2021, 3, 8–13. [Google Scholar] [CrossRef]
- Thalinger, B.; Deiner, K.; Harper, L.R.; Rees, H.C.; Blackman, R.C.; Sint, D.; Traugott, M.; Goldberg, C.S.; Bruce, K. A Validation Scale to Determine the Readiness of Environmental DNA Assays for Routine Species Monitoring. Environ. DNA 2021, 3, 823–836. [Google Scholar] [CrossRef]
- Cairns, D.K.; Benchetrit, J.; Bernatchez, L.; Bornarel, V.; Casselman, J.M.; Castonguay, M.; Charsley, A.R.; Dorow, M.; Drouineau, H.; Frankowski, J.; et al. Thirteen Novel Ideas and Underutilised Resources to Support Progress towards a Range-Wide American Eel Stock Assessment. Fish. Manag. Ecol. 2022, 29, 516–541. [Google Scholar] [CrossRef]
- Moyer, G.R.; Bartron, M.L.; Galbraith, H.S.; Grassi, J.; Rees, C.B. Development and Validation of Two Environmental DNA Assays for American Eel (Anguilla rostrata). Environ. DNA 2023, 5, 175–190. [Google Scholar] [CrossRef]
- Halvorsen, S.; Korslund, L.; Gustavsen, P.; Slettan, A. Environmental DNA Analysis Indicates That Migration Barriers Are Decreasing the Occurrence of European Eel (Anguilla anguilla) in Distance from the Sea. Glob. Ecol. Conserv. 2020, 24, e01245. [Google Scholar] [CrossRef]
- Halvorsen, S.; Korslund, L.; Mattingsdal, M.; Slettan, A. Estimating Number of European Eel (Anguilla anguilla) Individuals Using Environmental DNA and Haplotype Count in Small Rivers. Ecol. Evol. 2023, 13, e9785. [Google Scholar] [CrossRef]
- Cantera, I.; Decotte, J.B.; Dejean, T.; Murienne, J.; Vigouroux, R.; Valentini, A.; Brosse, S. Characterizing the Spatial Signal of Environmental DNA in River Systems Using a Community Ecology Approach. Mol. Ecol. Resour. 2022, 22, 1274–1283. [Google Scholar] [CrossRef]
- Rice, C.J.; Larson, E.R.; Taylor, C.A. Environmental DNA Detects a Rare Large River Crayfish but with Little Relation to Local Abundance. Freshw. Biol. 2018, 63, 443–455. [Google Scholar] [CrossRef]
- Jo, T.; Yamanaka, H. Meta-Analyses of Environmental DNA Downstream Transport and Deposition in Relation to Hydrogeography in Riverine Environments. Freshw. Biol. 2022, 67, 1333–1343. [Google Scholar] [CrossRef]
- Carraro, L.; Hartikainen, H.; Jokela, J.; Bertuzzo, E.; Rinaldo, A. Estimating Species Distribution and Abundance in River Networks Using Environmental DNA. Proc. Natl. Acad. Sci. USA 2018, 115, 11724–11729. [Google Scholar] [CrossRef] [PubMed]
- Zippin, C. The Removal Method of Population Estimation. J. Wildl. Manag. 1958, 22, 82–90. [Google Scholar] [CrossRef]
- Zippin, C. An Evaluation of the Removal Method of Estimating Animal Populations. Biometrics 1956, 12, 163–189. [Google Scholar] [CrossRef]
- Strahler, A.N. Quantitative Analysis of Watershed Geomorphology. Eos Trans. Am. Geophys. Union 1957, 38, 913–920. [Google Scholar] [CrossRef]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Paleontol. Stat. Softw. Stat 2013, 105, 1352–1357. [Google Scholar]
- Laramie, M.B.; Pilliod, D.S.; Goldberg, C.S. Characterizing the Distribution of an Endangered Salmonid Using Environmental DNA Analysis. Biol. Conserv. 2015, 183, 29–37. [Google Scholar] [CrossRef]
- Fernández, S.; Rodríguez, S.; Martínez, J.L.; Borrell, Y.J.; Ardura, A.; García-Vázquez, E. Evaluating Freshwater Macroinvertebrates from EDNA Metabarcoding: A River Nalón Case Study. PLoS ONE 2018, 13, e0201741. [Google Scholar] [CrossRef]
- Lobón-Cerviá, J.; Rincón, P. Variations in the Populationdynamics of the European Eel Anguilla anguilla (L.) along the Course of a Cantabrian River. Ecol. Freshw. Fish. 1995, 4, 17–27. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z. Estimation of Fish Biomass Using Environmental DNA. PLoS ONE 2012, 7, 3–10. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Laramie, M.B.; MacCoy, D.; Maclean, S. Integration of EDNA-Based Biological Monitoring within the U.S. Geological Survey’s National Streamgage Network. JAWRA J. Am. Water Resour. Assoc. 2019, 55, 1505–1518. [Google Scholar] [CrossRef]
- Itakura, H.; Wakiya, R.; Sakata, M.K.; Hsu, H.-Y.; Chen, S.-C.; Yang, C.-C.; Huang, Y.-C.; Han, Y.-S.; Yamamoto, S.; Minamoto, T.; et al. Estimations of Riverine Distribution, Abundance, and Biomass of Anguillid Eels in Japan and Taiwan Using Environmental DNA Analysis. Zool. Stud. 2020, 59, 17. [Google Scholar] [CrossRef]
- Knudsen, S.W.; Ebert, R.B.; Hesselsøe, M.; Kuntke, F.; Hassingboe, J.; Mortensen, P.B.; Thomsen, P.F.; Sigsgaard, E.E.; Hansen, B.K.; Nielsen, E.E.; et al. Species-Specific Detection and Quantification of Environmental DNA from Marine Fishes in the Baltic Sea. J. Exp. Mar. Biol. Ecol. 2019, 510, 31–45. [Google Scholar] [CrossRef]
- Muha, T.P. Using EDNA Metabarcoding to Monitor Changes in Fish Community Composition After Barrier Removal. Front. Ecol. Evol. 2021, 9, 28. [Google Scholar] [CrossRef]


| River | Location Name | Coordinates | Altitude (m) | Electrofishing Surface | Stream Order | Eel Biomass | Eel Number |
|---|---|---|---|---|---|---|---|
| Pigüeña | La Riera | 43°09′09.0″ N; 6°15′12.3″ O | 480 | 280 m2 | 3 | 1789 | 74 |
| Pigüeña | Santullano | 43°09′57.4″ N; 6°19′01.1″ O | 430 | 210 m2 | 2 | 418 | 17 |
| Pigüeña | Aguasmestas | 43°10′31.2″ N; 6°18′00.5″ O | 400 | 385 m2 | 4 | 1910 | 72 |
| Pigüeña | Belmonte P. Industrial | 43°17′17.6″ N; 6°13′16.94″ O | 200 | 455 m2 | 6 | 310 | 11 |
| Ponga | Las Mestas | 43°10′10.8″ N; 5°10′37.9″ O | 350 | 329 m2 | 4 | 290 | 6 |
| Location | Sample | Total eDNA Quantity | Universal Primers | Eel-Specific Primers |
|---|---|---|---|---|
| Aguasmestas | A1 | ND | 0 | 0 |
| A2 | ND | 0 | 0 | |
| A3 | 0.046 | 1 | 0 | |
| Belmonte | B1 | ND | 1 | 0 |
| B2 | ND | 0 | 1 | |
| B3 | 0.052 | 1 | 1 | |
| La Riera | LR1 | 0.032 | 1 | 1 |
| LR2 | 0.026 | 1 | 1 | |
| LR3 | ND | 1 | 0 | |
| Santullano | S1 | 0.158 | 1 | 0 |
| S2 | 0.114 | 1 | 1 | |
| S3 | 0.255 | 1 | 1 |
| Location | Sampling Replicate | PCR Replicate | CT | Adjusted Eel eDNA Quantity |
|---|---|---|---|---|
| Aguasmestas | A2 | A2.3 | 36.45 | 4.71 × 10−5 |
| A3 | A3.1 | 36.71 | 8.08 × 10−5 | |
| Belmonte | B1 | B1.1 | 36.58 | 8.84 × 10−5 |
| B2 | B2.1 | 34.37 | 1.85 × 10−4 | |
| B2.3 | 34.96 | 2.71 × 10−4 | ||
| B3 | B3.1 | 35.32 | 9.92 × 10−5 | |
| B3.2 | 34.22 | 2.06 × 10−4 | ||
| B3.3 | 34.07 | 5.02 × 10−4 | ||
| La Riera | LR1 | LR1.1 | 36.11 | 6.22 × 10−4 |
| LR1.3 | 37.13 | 6.05 × 10−5 | ||
| Santullano | S1 | S1.3 | 35.22 | 1.06 × 10−4 |
| S2 | S2.1 | 35.14 | 2.38 × 10−4 | |
| S3 | S3.2 | 35.25 | 1.00 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandez, S.; Gutiérrez, Á.; Deconinck, D.; Martinez, J.L.; Alvarez, A.; Marquez, I.; Machado-Schiaffino, G.; Garcia-Vazquez, E. Biomass Quantification of the Critically Endangered European eel from Running Waters Using Environmental DNA. Fishes 2023, 8, 279. https://doi.org/10.3390/fishes8060279
Fernandez S, Gutiérrez Á, Deconinck D, Martinez JL, Alvarez A, Marquez I, Machado-Schiaffino G, Garcia-Vazquez E. Biomass Quantification of the Critically Endangered European eel from Running Waters Using Environmental DNA. Fishes. 2023; 8(6):279. https://doi.org/10.3390/fishes8060279
Chicago/Turabian StyleFernandez, Sara, Álvaro Gutiérrez, Dumas Deconinck, Jose Luis Martinez, Almudena Alvarez, Isabel Marquez, Gonzalo Machado-Schiaffino, and Eva Garcia-Vazquez. 2023. "Biomass Quantification of the Critically Endangered European eel from Running Waters Using Environmental DNA" Fishes 8, no. 6: 279. https://doi.org/10.3390/fishes8060279





