Feature Importance for Human Epithelial (HEp-2) Cell Image Classification †
Abstract
:1. Introduction
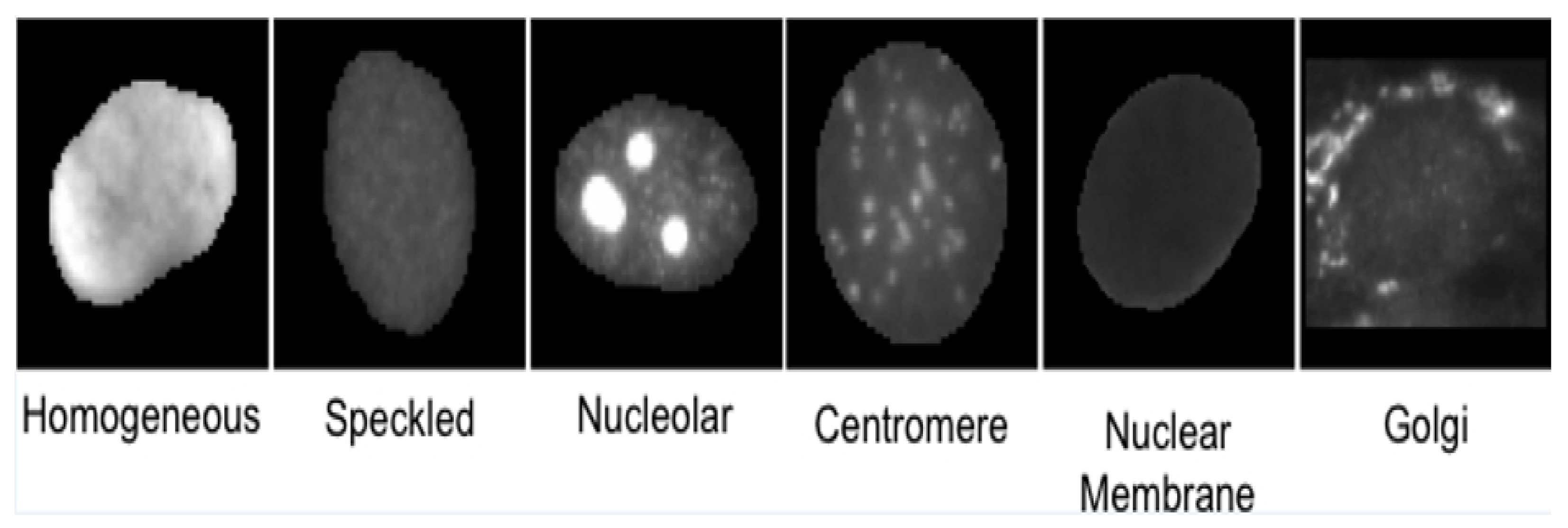
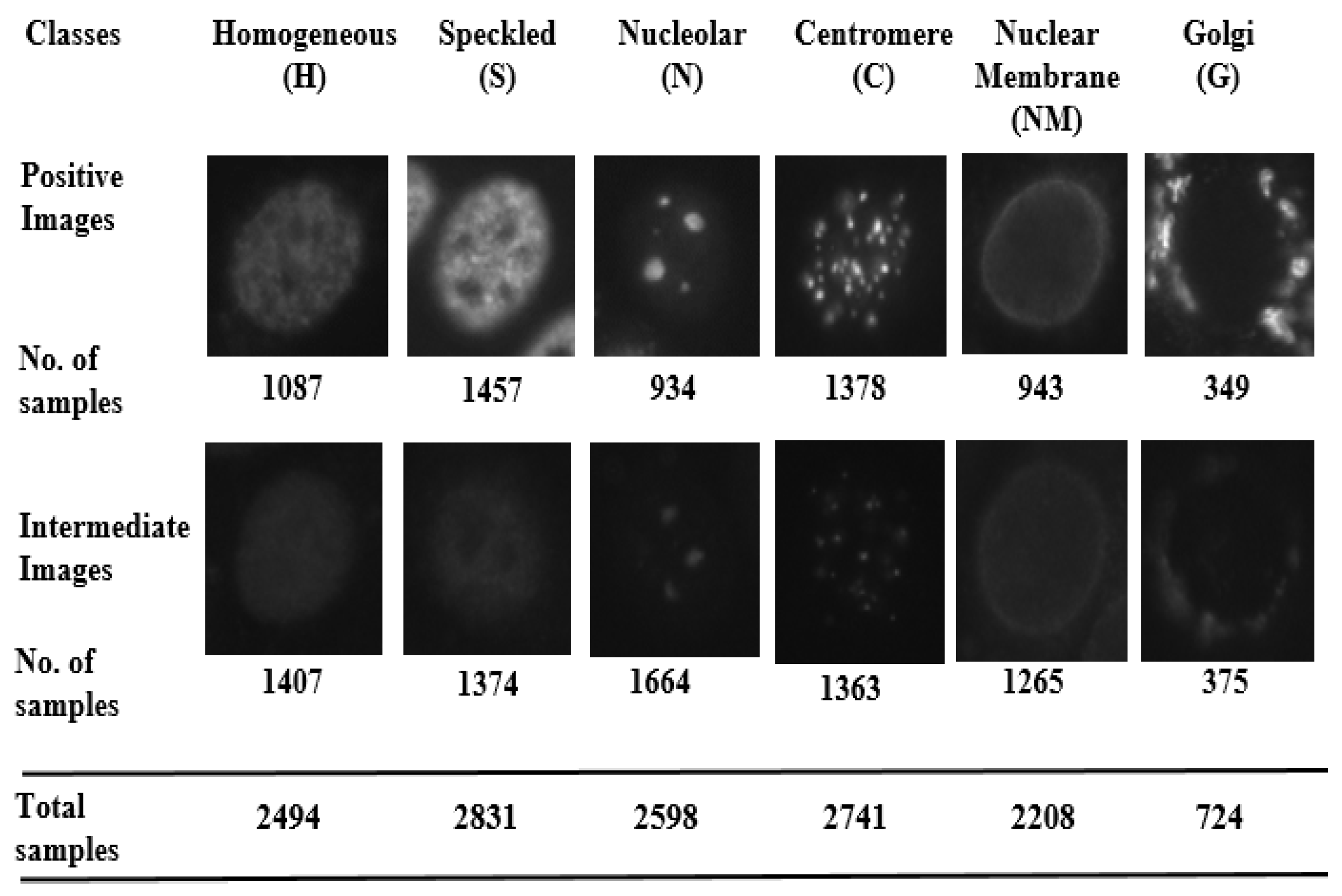
- Homogeneous: A uniformly diffused fluorescence covering the entire nucleoplasm sometimes accentuated in the nuclear periphery.
- Speckled: This pattern can be explained under two categories (which, however, are not separately labeled in the dataset)
- -
- Coarse speckled: Density distributed, various sized speckles, generally associated with larger speckles, throughout nucleoplasm of inter-phase cells; nucleoli are negative.
- -
- Fine speckled: Fine speckled staining in a uniform distributed, sometimes very dense so that an almost homogeneous pattern is attained; nucleoli may be positive and negative.
- Nucleolar: Characterized by clustered large granules in the nucleoli of inter phase cells which tend towards homogeneity, with less than six granules per cell.
- Centromere: Characterized by several discrete speckles (40–60) distributed throughout the inter phase nuclei.
- Nuclear Membrane: A smooth homogeneous ring-like fluorescence at the nuclear membrane.
- Golgi: Staining of polar organelle adjacent to and partly surrounding the nucleus, composed of irregular large granules. Nuclei and nucleoli are negative.
1.1. Feature Selection
- Filtering methods that use statistical properties of features to assign a score to each feature. Some examples are: information gain, chi-square test [17], fisher score, correlation coefficient, variance threshold etc. These methods are computationally efficient but do not consider the relationship between feature variables and response variable.
- Wrapper methods explore whole feature space to find an optimal subset by selecting features based on the classifier performance. These methods are computationally expensive due to the need to train and cross-validate model for each feature subset. Some examples are: recursive feature elimination, sequential feature selection [18] algorithms, genetic algorithms etc.
- Embedded methods perform feature selection as part of the learning procedure, and are generally specific for given classifier framework. Such approaches are more elegant than filtering methods, and computationally less intensive and less prone to over-fitting than wrapper methods. Some examples are: Random Forests [19] for Feature Ranking, Recursive Feature Elimination (RFE) with SVM, Adaboost for Feature Selection etc.
1.2. Scope of the Work
2. Related Work
3. Methodology
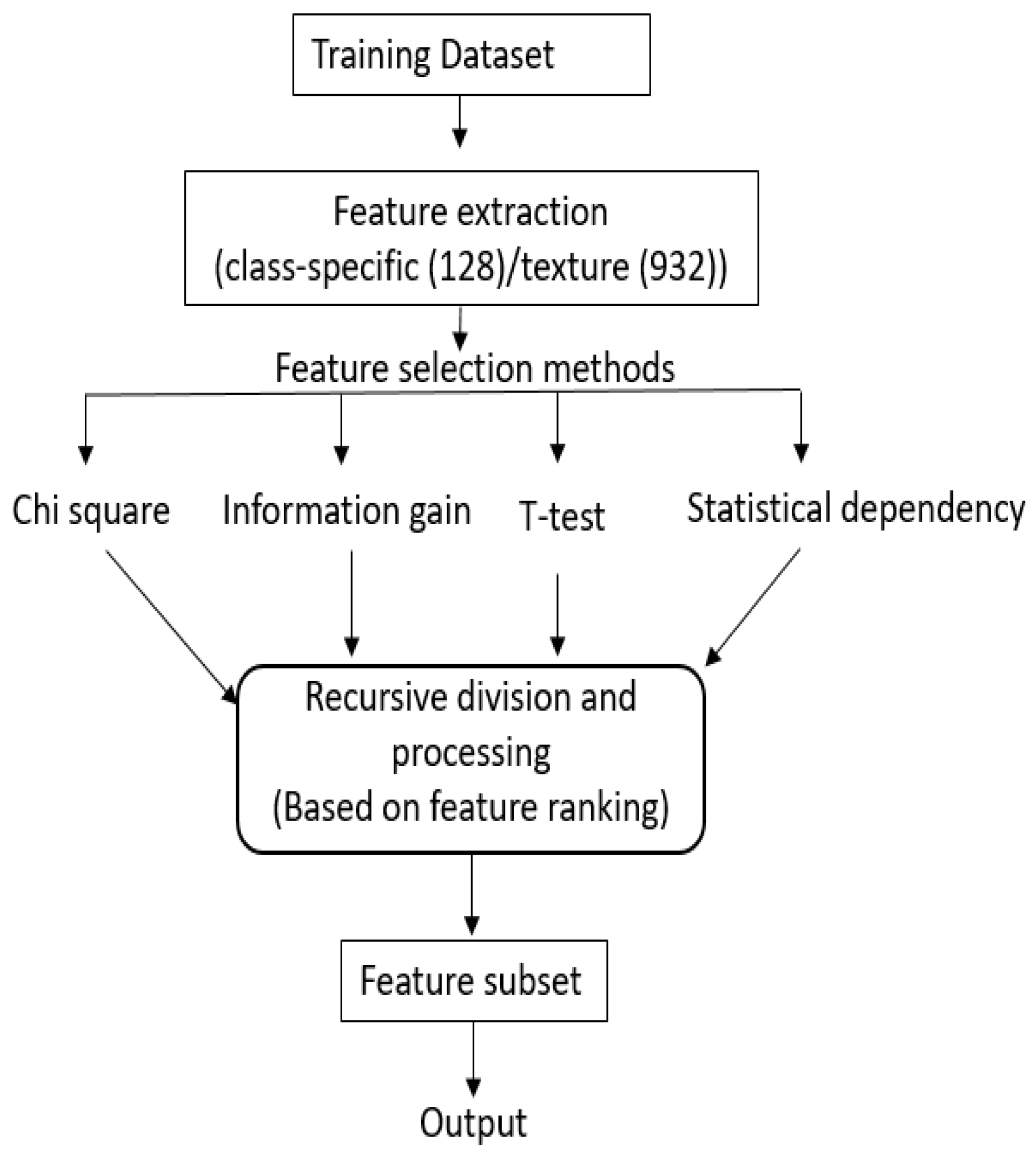
3.1. Filter Methods and Their Hybridization
3.1.1. Filter Methods
- Chi square [17]: The chi-square test is a statistical test that is applied to test the independence of two variables. In context of feature selection, it is analogous to hypothesis testing on distribution of class labels (target) and feature value. The greater the chi-score of a feature, the more independent that feature is from the class variable.
- t-test: t-test is a statistical, hypothesis where the statistic follows a Student distribution. After calculating the score of t-Statistic for each feature, scores are sorted in descending order in order to rank the features. t-test is usually applied between two groups of data. As our problem consists multiple groups (multiple classes), there are two ways by which t-test can be carried out. (1) Multiple paired t-test (2) one-way ANOVA (one-way analysis of variance). In this work, we used ANOVA, which uses p-value and F-statistics. In this work, we rank the features based on the F-statistics.
- Information gain (IG) [49]: computes how much information a feature gives about the class label. Typically, it is estimated as the difference between the class entropy and the conditional entropy in the presence of the feature.where C is the class variable, A is the attribute variable, and H( ) is the entropy. Features with higher IG score are ranked higher than features with lower scores.
- Statistical dependency (SD) [47]: measures the dependency between the features and their associated class labels. The larger the dependency, higher will be the feature score.
3.1.2. Hybridization Process
- We recursively divide the sorted feature set from each of the N filter methods, into halves based on ranking. For example, if total features are 128, than we first process 64 top features, followed by 32, 16, …, 8, 4, 2, 1.
- Processing the first half: (1) An initial selected feature set contains features that are common in first half of rank-sorted features from all N methods, (2) We then keep adding to this list the common features from among first halves of the rank-sorted list of methods, if the addition improves the accuracy. We follow this process with , , …, 2 methods. Note that at each stage, we consider common features from all combinations of , , …, 2 methods.
- Processing lower partitions: The above process is then carried out for the lower partitions and sub-partitions, recursively. Note that as one proceeds lower in the partitions, the number of features added in the final feature sets reduce, as many do not contribute to the increase in the accuracy. At each evaluation the improvement in accuracy is computed with a training and validation set by training a support vector machines (SVM).
| Algorithm 1 For finding feature subset using hybridization |
|
3.2. Wrapper Methods
- Sequential Forward Selection (SFS) [18]: It is an iterative method in which we start with an empty set. In each iteration, we keep adding the feature (one feature at a time) which best improves our model till an addition of a new variable does not improve the performance of the model. It is a greedy search algorithm, as it always excludes or includes the feature based of classification performance. The contribution of including or excluding a new feature is measured with respect to the set of previously chosen features using a hill climbing scheme in order to optimize the criterion function (classification performance).
- Random Subset Feature Selection (RSFS) [50]: In this method, in each step, random subset of features from all possible feature subset is chosen and estimated. The relevance of participating features keeps adjusting according to the model performance. As more iterations are performed, more relevant features obtain a higher score. and the quality of the feature set gradually improves. Unlike SFS, where the feature gain is estimated directly by excluding or including it from a existing set of features, in RSFS each feature is evaluated in terms of its average usefulness in the context of feature combinations. This method is not much susceptible to a locally optimal as SFS.
3.3. Embedded Methods and Feature Selection Procedure
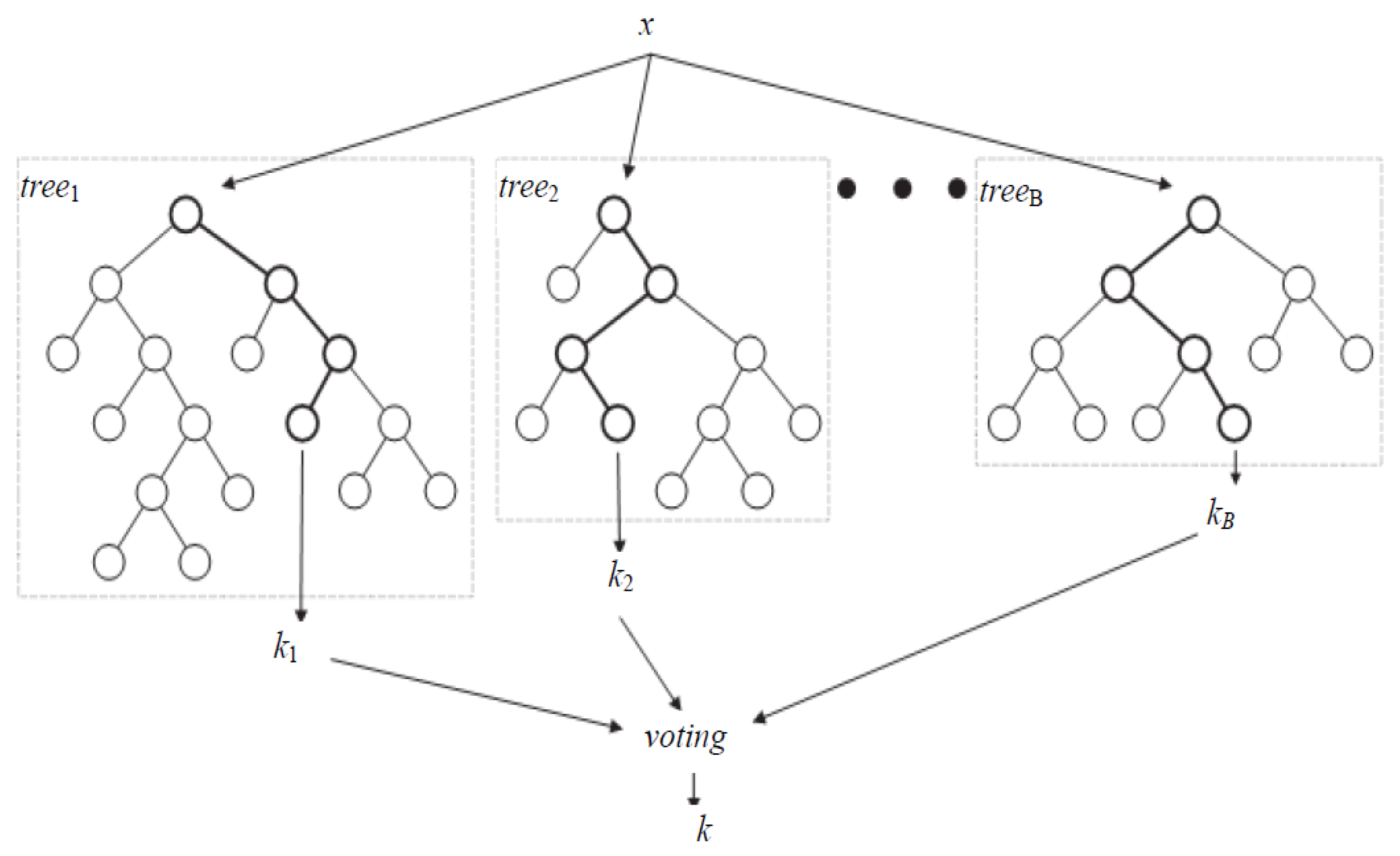
3.3.1. Random Forest
3.3.2. Random Uniform Forest (RUF)
3.3.3. Feature Selection Procedure
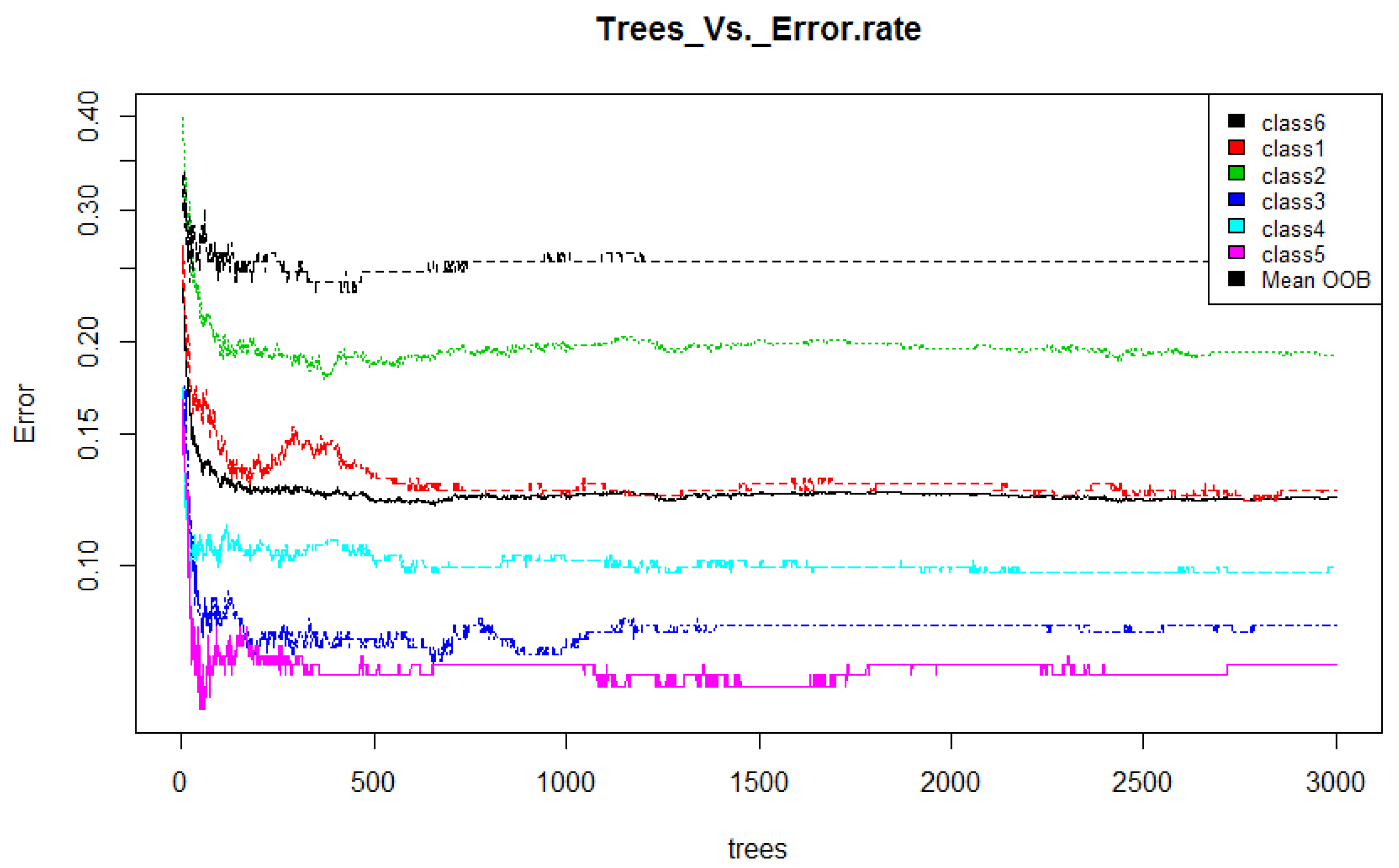
- Training: Random forest (RF) internally divides the training data into 67–33% ratio for each tree randomly, where 67% is used to train a model while 33% (out of bag) used for testing (cross-validation). The OOB error rate is calculated using this 33% data only. To automatically decide the value of RF parameters such as number of trees and number of features used at each tree, the OOB error rate is generally considered [52]. In this work, we only decide the number of trees. For the number of features at each tree, we employ typical default value (viz. square root of the total number of features). Figure 4 illustrates the variation in OOB error rate with the variation in the number of trees. There are seven lines in graph where six correspond to each class and the black line corresponds to mean of OOB error rate of all class. The point where the OOB error reduces negligibly could be considered as a good point to fix the value of number of trees [52].
- Feature selection: By using variable importance (VI) provided by RF (after cross-validation), a subset of good features is chosen to test the remaining 30% data. The procedure of selecting a good feature subset is given as follows:
- (a)
- Top ranked features (in terms of ) in sets of 10 (e.g., 10, 20, 30, …) are used for testing.
- (b)
- After some point, addition of more features do not increase the classification accuracy further.
- (c)
- As there is no significant improvement after this point, the features upto this point is considered as good feature subset. This feature subset gives the accuracy which is equal to the accuracy obtained using all features.
3.4. Unsupervised Feature Selection
- Infinite Feature Selection (InFS) [20]: It is graph-based method. Each feature is a node in the graph, a path is a selection of features, and the higher the centrality score, the most important (or most different) the feature.
- Regularized Discriminative Feature Selection (UDFS) [21]: It is a L2,1-norm regularized discriminative feature selection method which simultaneously exploits discriminative information and feature correlations. It selects most discriminative feature subset from the whole feature set in batch mode.
- Multi Cluster feature selection (MCFS) [22]: It selects the features for those e multi-cluster structure of the data can be best preserved. Having used, spectral analysis it suggests a way to measure the correlations between different features without label information. Hence, it can well handle the data with multiple cluster structure.
4. Dataset and Feature Extraction
4.1. Dataset
4.2. Feature Extraction
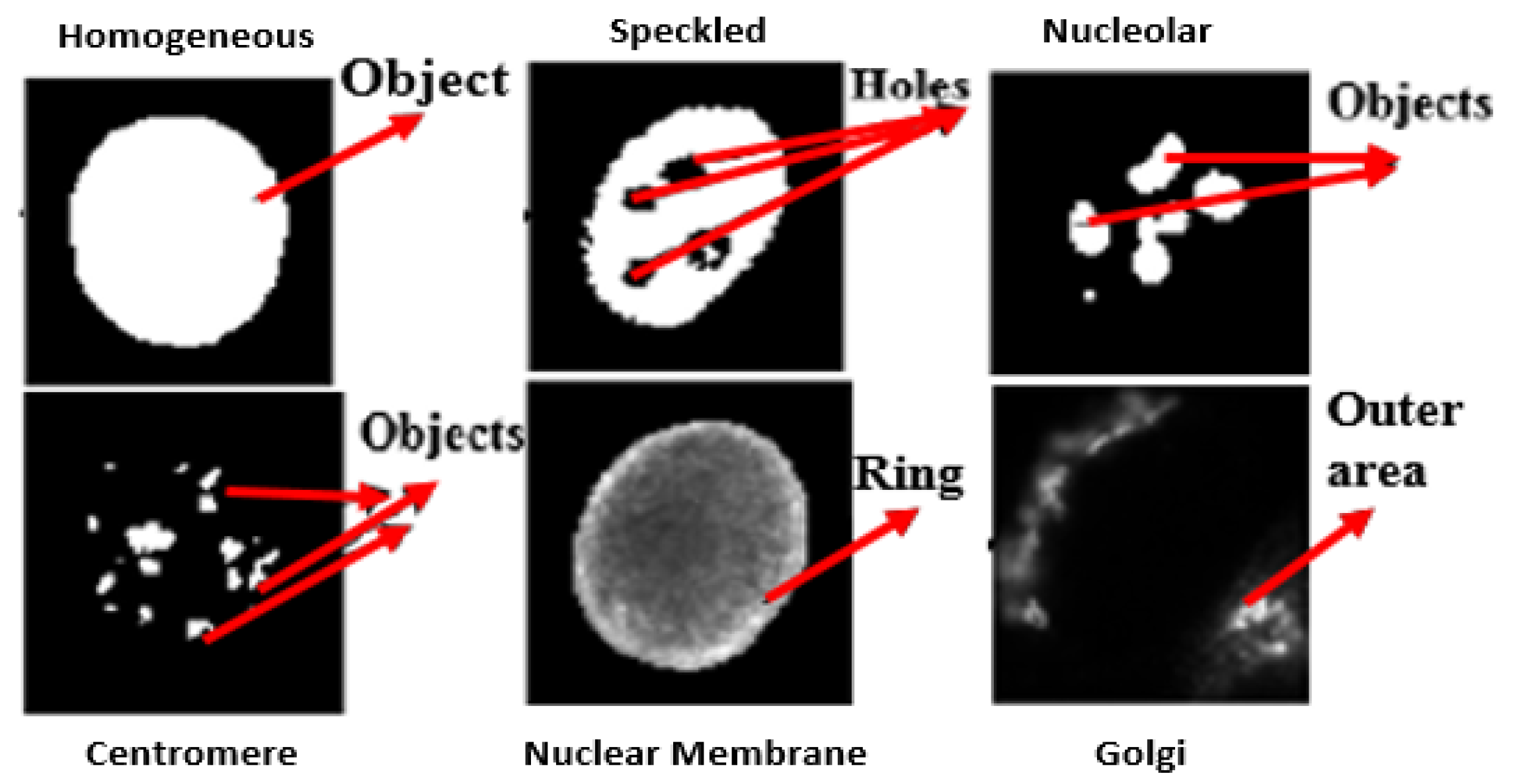
- Class-specific features: Motivated by expert knowledge [9] which characterizes each class by giving some unique morphological traits, we define features based on such traits. As the location of these traits in each class may be different, features are extracted from specific regions of interest (ROI) for each particular class, computed using the mask images. Figure 6 shows some of the unique traits of each class. For example, in NM class useful information can be found in a ring which is centered on the boundary. Thus, utilizing such visually observed traits following features are extracted for the NM class:
- 1
- Boundary area ratio (BAR): It is a area ratio in boundary ring mask.
- 2
- Inner area ratio (IAR): It is a area ratio in inner mask.
- 3
- Eroded area connected component (EACC): It gives the number of white pixels in inner mask.
Similarly, for the Centromere class the class specific features defined are:- 1
- Maximum object Area (MOA): Area of object having a maximum size.
- 2
- Average object Area (AOA): Average area of objects.
- 3
- Connected Component (CC): Total number of objects in the cell.
Here we only mention a list of these features in Table 1, but and we refer the reader to our previous work [13] which contains a more elaborate discussion. These features, involve simple image processing operations such as scaler image enhancement, thresholding, connected components. and morphological operation. Our earlier work [13] explains the features in more detail. These feature are scalar-valued, simple, efficient, and more interpretable. As listed in Table 1, the total feature definitions which are extracted from all classes is 18, and using various combination of threshold and enhancement parameters, a total 128 features are obtained. - Traditional scalar texture features [14]: These include features which include morphology descriptors (like Number of objects, Area, Area of the convex hull, Eccentricity, Euler number, Perimeter), and texture descriptors (e.g., Intensity, Standard deviation, Entropy, Range, GLCM) are extracted at 20 intensity thresholds equally spaced from its minimum to its maximum intensity. Again for a detailed description about these features, one can refer [14]. However, in [14], these features were applied on much smaller dataset. Considering various parameter variations, in this case, a much larger set of 932 scalar-valued features is obtained.
5. Results and Discussion
5.1. Training and Testing Protocols
5.2. Evaluation Metrics
- : Highest accuracy with respect to the number of selected features.
- : Number of features that yield the highest accuracy.
- : Accuracy considering all features.
- : Number of features that match the accuracy considering all features.
5.3. Results: Protocol 1
5.3.1. Filter Methods
5.3.2. Hybridization
5.3.3. Wrapper Methods
5.3.4. Embedded Methods
5.3.5. Unsupervised Feature Selection Methods
5.4. Results: Protocol 2
5.5. Comparison among All Feature Selection Methods
5.6. Performance Comparison with Contemporary Methods
5.7. Computational Time Analysis
6. Conclusions
Author Contributions
Conflicts of Interest
References
- Sack, U.; Conrad, K.; Csernok, E.; Frank, I.; Hiepe, F.; Krieger, T.; Kromminga, A.; Von Landenberg, P.; Messer, G.; Witte, T.; et al. Autoantibody Detection Using Indirect Immunofluorescence on HEp-2 Cells. Ann. N. Y. Acad. Sci. 2009, 1173, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Friou, G.J.; Finch, S.C.; Detre, K.D.; Santarsiero, C. Interaction of nuclei and globulin from lupus erythematosis serum demonstrated with fluorescent antibody. J. Immunol. 1958, 80, 324–329. [Google Scholar] [PubMed]
- Foggia, P.; Percannella, G.; Soda, P.; Vento, M. Benchmarking HEp-2 cells classification methods. IEEE Trans. Med. Imaging 2013, 32, 1878–1889. [Google Scholar] [CrossRef] [PubMed]
- Wiik, A.S.; Høier-Madsen, M.; Forslid, J.; Charles, P.; Meyrowitsch, J. Antinuclear antibodies: A contemporary nomenclature using HEp-2 cells. J. Autoimmun. 2010, 35, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Bhatia, A.; Minz, R.W. Antinuclear antibodies and their detection methods in diagnosis of connective tissue diseases: A journey revisited. Diagn. Pathol. 2009, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hobson, P.; Lovell, B.C.; Percannella, G.; Vento, M.; Wiliem, A. Benchmarking human epithelial type 2 interphase cells classification methods on a very large dataset. Artif. Intell. Med. 2015, 65, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Hobson, P.; Percannella, G.; Vento, M.; Wiliem, A. Competition on cells classification by fluorescent image analysis. In Proceedings of the 20th IEEE International Conference on Image Processing (ICIP), Melbourne, Australia, 15–18 September 2013; pp. 2–9. [Google Scholar]
- Bradwell, A.; Hughes, R.S.; Harden, E. Atlas of Hep-2 Patterns and Laboratory Techniques; Binding Site: Birmingham, UK, 1995. [Google Scholar]
- Foggia, P.; Percannella, G.; Soda, P.; Vento, M. Early experiences in mitotic cells recognition on HEp-2 slides. In Proceedings of the 2010 IEEE 23rd International Symposium on Computer-Based Medical Systems (CBMS), Perth, Australia, 12–15 October 2010; pp. 38–43. [Google Scholar]
- Iannello, G.; Percannella, G.; Soda, P.; Vento, M. Mitotic cells recognition in HEp-2 images. Pattern Recognit. Lett. 2014, 45, 136–144. [Google Scholar] [CrossRef]
- Ensafi, S.; Lu, S.; Kassim, A.A.; Tan, C.L. A bag of words based approach for classification of hep-2 cell images. In Proceedings of the 2014 1st Workshop on Pattern Recognition Techniques for Indirect Immunofluorescence Images (I3A), Stockholm, Sweden, 24 August 2014; pp. 29–32. [Google Scholar]
- Stoklasa, R.; Majtner, T.; Svoboda, D. Efficient k-NN based HEp-2 cells classifier. Pattern Recognit. 2014, 47, 2409–2418. [Google Scholar] [CrossRef]
- Gupta, V.; Gupta, K.; Bhavsar, A.; Sao, A.K. Hierarchical classification of HEp-2 cell images using class-specific features. In Proceedings of the European Workshop on Visual Information Processing (EUVIP 2016), Marseille, France, 25–27 October 2016. [Google Scholar]
- Strandmark, P.; Ulén, J.; Kahl, F. Hep-2 staining pattern classification. In Proceedings of the 2012 21st International Conference on Pattern Recognition (ICPR), Tsukuba, Japan, 11–15 November 2012; pp. 33–36. [Google Scholar]
- Blum, A.L.; Langley, P. Selection of relevant features and examples in machine learning. Artif. Intell. 1997, 97, 245–271. [Google Scholar] [CrossRef]
- Reunanen, J. Overfitting in making comparisons between variable selection methods. J. Mach. Learn. Res. 2003, 3, 1371–1382. [Google Scholar]
- Liu, H.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the Seventh International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; pp. 388–391. [Google Scholar]
- Whitney, A.W. A direct method of nonparametric measurement selection. IEEE Trans. Comput. 1971, 100, 1100–1103. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Roffo, G.; Melzi, S.; Cristani, M. Infinite Feature Selection. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Washington, DC, USA, 7–13 December 2015; pp. 4202–4210. [Google Scholar]
- Yang, Y.; Shen, H.T.; Ma, Z.; Huang, Z.; Zhou, X. L2,1-norm Regularized Discriminative Feature Selection for Unsupervised Learning. In Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence (IJCAI’11)—Volume Volume Two, Barcelona, Spain, 16–22 July 2011; pp. 1589–1594. [Google Scholar]
- Cai, D.; Zhang, C.; He, X. Unsupervised feature selection for multi-cluster data. In Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Washington, DC, USA, 24–28 July 2010; ACM: New York, NY, USA, 2010; pp. 333–342. [Google Scholar]
- Yang, Y.H.; Xiao, Y.; Segal, M.R. Identifying differentially expressed genes from microarray experiments via statistic synthesis. Bioinformatics 2004, 21, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Ciss, S. Variable Importance in Random Uniform Forests. 2015. [Google Scholar]
- Gupta, V.; Bhavsar, A. Random Forest-Based Feature Importance for HEp-2 Cell Image Classification. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis, dinburgh, UK, 11–13 July 2017; Communications in Computer and Information Science (CCIS). Springer: Cham, Switzerland, 2017; Volume 723, pp. 922–934. [Google Scholar]
- Perner, P.; Perner, H.; Müller, B. Mining knowledge for HEp-2 cell image classification. Artif. Intell. Med. 2002, 26, 161–173. [Google Scholar] [CrossRef]
- Huang, Y.C.; Hsieh, T.Y.; Chang, C.Y.; Cheng, W.T.; Lin, Y.C.; Huang, Y.L. HEp-2 cell images classification based on textural and statistic features using self-organizing map. In Proceedings of the Asian Conference on Intelligent Information and Database Systems, Kaohsiung, Taiwan, 19–21 March 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 529–538. [Google Scholar]
- Hsieh, T.Y.; Huang, Y.C.; Chung, C.W.; Huang, Y.L. HEp-2 cell classification in indirect immunofluorescence images. In Proceedings of the 7th International Conference on Information, Communications and Signal Processing ( ICICS), Macau, China, 8–10 December 2009; pp. 1–4. [Google Scholar]
- Foggia, P.; Percannella, G.; Saggese, A.; Vento, M. Pattern recognition in stained HEp-2 cells: Where are we now? Pattern Recognit. 2014, 47, 2305–2314. [Google Scholar] [CrossRef]
- Prasath, V.; Kassim, Y.; Oraibi, Z.A.; Guiriec, J.B.; Hafiane, A.; Seetharaman, G.; Palaniappan, K. HEp-2 cell classification and segmentation using motif texture patterns and spatial features with random forests. In Proceedings of the 23th International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016. [Google Scholar]
- Li, B.H.; Zhang, J.; Zheng, W.S. HEp-2 cells staining patterns classification via wavelet scattering network and random forest. In Proceedings of the 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR), Kuala Lumpur, Malaysia, 3–6 November 2015; pp. 406–410. [Google Scholar]
- Agrawal, P.; Vatsa, M.; Singh, R. HEp-2 cell image classification: A comparative analysis. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Nagoya, Japan, 22 September 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 195–202. [Google Scholar]
- Manivannan, S.; Li, W.; Akbar, S.; Wang, R.; Zhang, J.; McKenna, S.J. An automated pattern recognition system for classifying indirect immunofluorescence images of HEp-2 cells and specimens. Pattern Recognit. 2016, 51, 12–26. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, L.; Zhou, L.; Zhang, J. Hep-2 cell image classification with deep convolutional neural networks. arXiv, 2015; arXiv:1504.02531. [Google Scholar]
- Hobson, P.; Lovell, B.C.; Percannella, G.; Saggese, A.; Vento, M.; Wiliem, A. HEp-2 staining pattern recognition at cell and specimen levels: Datasets, algorithms and results. Pattern Recognit. Lett. 2016, 82, 12–22. [Google Scholar] [CrossRef]
- Hobson, P.; Lovell, B.C.; Percannella, G.; Saggese, A.; Vento, M.; Wiliem, A. Computer aided diagnosis for anti-nuclear antibodies HEp-2 images: progress and challenges. Pattern Recognit. Lett. 2016, 82, 3–11. [Google Scholar] [CrossRef]
- Gragnaniello, D.; Sansone, C.; Verdoliva, L. Biologically-inspired dense local descriptor for indirect immunofluorescence image classification. In Proceedings of the 2014 1st Workshop on Pattern Recognition Techniques for Indirect Immunofluorescence Images (I3A), Stockholm, Sweden, 24 August 2014; pp. 1–5. [Google Scholar]
- Moorthy, K.; Mohamad, M.S. Random forest for gene selection and microarray data classification. In Knowledge Technology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 174–183. [Google Scholar]
- Paul, A.; Dey, A.; Mukherjee, D.P.; Sivaswamy, J.; Tourani, V. Regenerative Random Forest with Automatic Feature Selection to Detect Mitosis in Histopathological Breast Cancer Images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 94–102. [Google Scholar]
- Nguyen, C.; Wang, Y.; Nguyen, H.N. Random forest classifier combined with feature selection for breast cancer diagnosis and prognostic. J. Biomed. Sci. Eng. 2013, 6, 551–560. [Google Scholar] [CrossRef]
- Hariharan, S.; Tirodkar, S.; De, S.; Bhattacharya, A. Variable importance and random forest classification using RADARSAT-2 PolSAR data. In Proceedings of the 2014 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Quebec City, QC, Canada, 13–18 July 2014; pp. 1210–1213. [Google Scholar]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Tsai, C.F.; Hsiao, Y.C. Combining multiple feature selection methods for stock prediction: Union, intersection, and multi-intersection approaches. Decis. Support Syst. 2010, 50, 258–269. [Google Scholar] [CrossRef]
- Al-Thubaity, A.; Abanumay, N.; Al-Jerayyed, S.; Alrukban, A.; Mannaa, Z. The effect of combining different feature selection methods on arabic text classification. In Proceedings of the 2013 14th ACIS International Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing (SNPD), Honolulu, HI, USA, 1–3 July 2013; pp. 211–216. [Google Scholar]
- Rajab, K.D. New Hybrid Features Selection Method: A Case Study on Websites Phishing. Secur. Commun. Netw. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Pohjalainen, J.; Räsänen, O.; Kadioglu, S. Feature selection methods and their combinations in high- dimensional classification of speaker likability, intelligibility and personality traits. Comput. Speech Lang. 2015, 29, 145–171. [Google Scholar] [CrossRef]
- Doan, S.; Horiguchi, S. An efficient feature selection using multi-criteria in text categorization. In Proceedings of the 2004 Fourth International Conference on Hybrid Intelligent Systems (HIS’04), Kitakyushu, Japan, 5–8 December 2004; pp. 86–91. [Google Scholar]
- Cover, T.M.; Thomas, J.A. Elements of Information Theory; Wiley: Hoboken, NJ, USA, 1991. [Google Scholar]
- Räsänen, O.; Pohjalainen, J. Random subset feature selection in automatic recognition of developmental disorders, affective states, and level of conflict from speech. In Proceedings of the INTERSPEECH, Lyon, France, 25–29 August 2013; pp. 210–214. [Google Scholar]
- Strobl, C.; Zeileis, A. Danger: High power!—Exploring the Statistical Properties of a Test for Random Forest Variable Importance. 2008. Available online: https://epub.ub.uni-muenchen.de/2111/ (accessed on 14 February 2018).
- Segal, M.R. Machine Learning Benchmarks and Random Forest Regression; Center for Bioinformatics & Molecular Biostatistics: California, CA, USA, 2004. [Google Scholar]
- Liu, H.; Motoda, H. Feature Selection for Knowledge Discovery and Data Mining; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 454. [Google Scholar]
| Classes | Class-Specific Features | ||
|---|---|---|---|
| Homogeneous | Maximum object area (MOA), Area of connected component (ACC), Maximum perimeter (MP) | ||
| Speckled | Hole area (HA), Hole number (HN), Euler number (EN) | ||
| Nuleolar | Maximum object area (MOA), Average object area (AOA), Connected component (CC) | ||
| Centromere | Maximum object area (MOA), Average object area (AOA), Connected component (CC) | ||
| Nuclear Membrane | Boundary area ratio (BAR), Inner area ratio (IAR), Eroded area connected component (EACC) | ||
| Golgi | Outer area ratio (OAR), Average object distance (AOD), Eroded area, connected component (EACC) | ||
| Class-Specific Features (Total Features: 128) | Texture Features (Total Features: 932) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Methods/Trials | Chi Square | t-Test | Information Gain | Statistical Dependency | Chi Square | t-Test | Information Gain | Statistical Dependency | |
| Average (Validation) | Ah | 98.43 | 98.47 | 97.98 | 98.43 | 95.14 | 94.97 | 95.14 | 94.93 |
| Nh | 98 | 98 | 93 | 98 | 756 | 812 | 702 | 863 | |
| As | 98.23 | 98.12 | 97.57 | 98.23 | 94.72 | 94.72 | 94.73 | 94.72 | |
| Ns | 91 | 85 | 84 | 91 | 730 | 693 | 666 | 838 | |
| Testing | Ah | 98.16 | 98.16 | 97.44 | 98.23 | 94.74 | 94.50 | 95.05 | 94.26 |
| Testing | As | 98.10 | 97.41 | 97.03 | 98.13 | 94.35 | 94.17 | 95.00 | 94.20 |
| Hybridization | ||||||||
|---|---|---|---|---|---|---|---|---|
| Class-Specific Features (Total Features: 128) | Texture Features (Total Features: 932) | |||||||
| Trail 1 | Trail 2 | Trail 3 | Trail 4 | Trail 1 | Trail 2 | Trail 3 | Trail 4 | |
| Ah | 98.33 | 98.65 | 97.98 | 98.62 | 95.09 | 95.48 | 95.31 | 95.90 |
| Nh | 86 | 91 | 85 | 95 | 521 | 536 | 546 | 585 |
| As | 98.33 | 98.65 | 97.98 | 98.62 | 95.04 | 95.31 | 95.31 | 95.90 |
| Ns | 86 | 91 | 85 | 95 | 478 | 520 | 546 | 585 |
| Average Results (Validation) | ||||||||
| Ah | 98.39 | 95.45 | ||||||
| Nh | 89 | 547 | ||||||
| As | 98.39 | 95.39 | ||||||
| Ns | 89 | 532 | ||||||
| Testing | ||||||||
| Ah | 98.28 | 94.90 | ||||||
| As | 98.28 | 94.90 | ||||||
| Class-Specific Features (Total Features: 128) | Texture Features (Total Features: 932) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Methods | Wrapper | Embedded | Wrapper | Embedded | |||||
| SFS | RSFS | RF | RUF | SFS | RSFS | RF | RUF | ||
| Average (Validation) | Ah | 98.17 | 96.92 | 97.44 | 97.78 | 90.57 | 94.44 | 94.29 | 95.25 |
| Nh | 87 | 30 | 66 | 60 | 35 | 133 | 202 | 258 | |
| As | - | - | 97.22 | 97.66 | - | - | 93.75 | 94.87 | |
| Ns | - | - | 41 | 32 | - | - | 60 | 154 | |
| Testing | Ah | 97.88 | 96.78 | 97.19 | 97.55 | 88.14 | 90.62 | 94.18 | 95.13 |
| Testing | As | - | - | 97.13 | 97.36 | - | - | 93.60 | 94.84 |
| Class-Specific (128) | Texture (932) | |||||
|---|---|---|---|---|---|---|
| Average | MCFS | InFS | UDFS | MCFS | InFS | UDFS |
| Ah | 98.35 | 98.06 | 98.15 | 94.88 | 94.81 | 94.80 |
| Nh | 108 | 118 | 99 | 918 | 811 | 853 |
| As | 98.03 | 98.03 | 93.03 | 90.39 | 90.20 | 89.26 |
| Ns | 91 | 115 | 89 | 321 | 323 | 291 |
| Testing (Ah) | 98.17 | 97.95 | 98.10 | 94.75 | 94.69 | 94.60 |
| Testing (As) | 97.94 | 97.94 | 98.05 | 90.63 | 89.69 | 89.27 |
| Class-Specific features (Total Features: 128) | |||||||
| Filter Methods | Embedded | ||||||
| Average | Chi Square | t-Test | IG | SD | Hybrid | RF | RUF |
| Ah | 98.73 | 98.73 | 98.72 | 98.65 | 98.65 | 97.51 | 97.87 |
| Nh | 95 | 113 | 108 | 118 | 85 | 66 | 54 |
| As | 98.64 | 98.62 | 98.62 | 98.62 | 98.65 | 97.32 | 97.77 |
| Ns | 86 | 105 | 97 | 118 | 85 | 40 | 38 |
| Testing (Ah) | 98.66 | 98.49 | 98.51 | 98.54 | 98.59 | 97.51 | 97.78 |
| Testing (As) | 98.51 | 98.10 | 98.42 | 98.40 | 98.59 | 97.26 | 97.60 |
| Texture Features (Total Features: 932) | |||||||
| Ah | 95.82 | 96.61 | 95.83 | 95.95 | 95.90 | 94.67 | 94.93 |
| Nh | 810 | 847 | 750 | 791 | 465 | 250 | 330 |
| As | 95.47 | 95.47 | 95.47 | 95.47 | 95.90 | 93.89 | 94.60 |
| Ns | 742 | 798 | 465 | 675 | 465 | 80 | 120 |
| Testing (Ah) | 96.00 | 95.76 | 95.51 | 96.00 | 95.51 | 94.21 | 95.11 |
| Testing (As) | 95.92 | 95.71 | 95.51 | 95.91 | 95.51 | 93.01 | 94.91 |
| Class-Specific (128) | Texture (932) | |||||
|---|---|---|---|---|---|---|
| Average | MCFS | InFS | UDFS | MCFS | InFS | UDFS |
| Ah | 98.80 | 98.67 | 98.71 | 95.70 | 95.58 | 95.60 |
| Nh | 97 | 116 | 117 | 905 | 797 | 816 |
| As | 98.62 | 98.62 | 98.61 | 90.16 | 90.55 | 89.95 |
| Ns | 90 | 113 | 121 | 284 | 280 | 284 |
| Testing (Ah) | 98.66 | 98.49 | 98.46 | 95.77 | 95.83 | 95.78 |
| Testing (As) | 98.63 | 98.21 | 98.32 | 90.34 | 90.75 | 90.26 |
| Class-Specific Features (Total Features: 128) | |||||||||
| Filter | Wrapper | Embeded | |||||||
| Average | Chi Square | t-Test | Information Gain | Statistical Dependency | Hybrid | SFS | RSFS | RF | RUF |
| Ah | 98.43 | 98.47 | 97.98 | 98.43 | 98.39 | 98.17 | 96.92 | 97.44 | 97.78 |
| Nh | 98 | 98 | 93 | 98 | 89 | 87 | 30 | 66 | 60 |
| As | 97.57 | 98.12 | 98.23 | 98.03 | 98.39 | - | - | 97.22 | 97.66 |
| Ns | 91 | 85 | 84 | 91 | 89 | - | - | 41 | 32 |
| Testing (Ah) | 98.16 | 98.16 | 97.44 | 98.23 | 98.28 | 97.88 | 96.78 | 97.19 | 97.55 |
| Testing (As) | 98.10 | 97.41 | 97.03 | 98.13 | 98.28 | 97.88 | 96.78 | 97.13 | 97.36 |
| Texture Features (Total Features: 932) | |||||||||
| Ah | 95.14 | 94.97 | 95.14 | 94.93 | 95.45 | 90.57 | 94.44 | 94.29 | 95.25 |
| Nh | 756 | 812 | 702 | 863 | 547 | 35 | 133 | 202 | 258 |
| As | 94.72 | 94.72 | 94.73 | 94.72 | 95.39 | - | - | 93.75 | 94.87 |
| Ns | 730 | 693 | 666 | 838 | 532 | - | - | 60 | 154 |
| Testing (Ah) | 94.74 | 94.50 | 95.05 | 94.26 | 94.90 | 88.14 | 90.62 | 94.18 | 95.13 |
| Testing (As) | 94.35 | 94.06 | 95.00 | 94.20 | 94.90 | 88.14 | 90.62 | 93.60 | 94.84 |
| Methods | Experimental-Setup | Metrics Evaluation | Nh (Number of Features) | ||
|---|---|---|---|---|---|
| MCA (%) | FP (%) | OA (%) | |||
| Prasath et al. [30] | 5-fold cross-validation (Training: 80%, Testing: 20%) | 94.29 | NA | NA | 552 |
| Bran et al. [31] | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 89.79 | 10.33 | 90.59 | NA |
| Praful et al. [32] | 10-fold cross-validation | NA | NA | 92.85 ± 0.63 | 211 |
| Shahab et al. [11] | 7-fold cross-validation | 94.9 | 5.09 | 94.48 | NA |
| Siyamalan et al. [33] | 2-fold cross-validation (each repeated 10 times) | 92.58 (95.21) | 7.38 (4.8) | NA | NA |
| Zhimin et al. [34] | Data-augmentation (Training: 64%, validation: 16%, Testing: 20%) | 88.58 (96.76) | NA | 89.04 (97.24) | NA |
| Supervised Feature Selection | |||||
| Proposed (Filter: Chi): class-specific | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 98.62 ± 0.37 | 1.70 ± 0.56 | 98.66 ± 0.30 | 95 |
| Proposed (Embedded: RUF): class-specific | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 97.46 ± 0.55 | 0.49 ± 0.06 | 97.73 ± 0.32 | 54 |
| Proposed (Filter: Chi): Texture | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 95.23 ± 0.81 | 4.19 ± 0.74 | 96.00 ± 0.52 | 810 |
| Proposed (Embedded: RUF): Texture | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 93.92 ± 0.92 | 1.09 ± 0.12 | 95.11 ± 0.55 | 330 |
| Proposed (Filter: SD): class-specific | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 98.12 ± 0.32 | 2.19 ± 0.55 | 98.23 ± 0.38 | 98 |
| Proposed (Wrapper: SFS): class-specific | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 97.88 ± 0.39 | 2.44 ± 0.22 | 97.73 ± 0.37 | 87 |
| Proposed (Embedded: RUF): class-specific | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 97.55±0.52 | 0.51 ± 0.07 | 97.36 ± 0.45 | 60 |
| Proposed (Filter: IG): Texture | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 93.79 ± 0.23 | 5.66 ± 0.61 | 95.05 ± 0.43 | 702 |
| Proposed (Wrapper: RSFS): Texture | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 90.62 ± 0.55 | 9.74 ± 0.56 | 89.15 ± 0.34 | 133 |
| Proposed (Embedded: RUF): Texture | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 95.13 ± 0.67 | 1.06 ± 0.17 | 94.06 ± 0.81 | 258 |
| Unsupervised Feature Selection | |||||
| Proposed: Class-specific | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 98.62 ± 0.22 | 1.67 ± 0.53 | 98.66 ± 0.32 | 97 |
| Proposed: Texture | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 94.95 ± 0.41 | 4.39 ± 0.68 | 95.83 ± 0.74 | 905 |
| Proposed: Class-specific | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 97.88 ± 0.18 | 2.18 ± 0.53 | 98.14 ± 0.18 | 108 |
| Proposed: Texture | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 93.29 ± 0.32 | 5.90 ± 0.16 | 94.75 ± 0.07 | 914 |
| Hybridization | |||||
| Proposed (hybrid): class-specific | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 98.48 ± 0.35 | 1.78 ± 0.67 | 98.59 ± 0.31 | 85 |
| Hybrid (exist): class-specific | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 98.59 ± 0.38 | 1.68 ± 0.56 | 98.65 ± 0.30 | 99 |
| Proposed (hybrid): Texture | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 94.72 ± 1.04 | 4.70 ± 0.77 | 95.51 ± 0.54 | 465 |
| Hybrid (exist): Texture | 5-fold cross-validation (Training: 64%, validation: 16%, Testing: 20%) | 95.11 ± 0.70 | 4.29 ± 0.42 | 95.90 ± 0.71 | 819 |
| Proposed (hybrid): class-specific | Training-40%, validation-30%, Testing-30% Repeated for four random trails | 98.15 ± 0.48 | 2.09 ± 0.43 | 98.23 ± 0.23 | 89 |
| Hybrid (exist): class-specific | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 98.21 ± 0.32 | 2.10 ± 0.57 | 98.29 ± 0.29 | 94 |
| Proposed (hybrid): Texture | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 93.84 ± 0.76 | 5.44 ± 0.34 | 94..90 ± 0.43 | 547 |
| Hybrid (exist): Texture | Training: 40%, validation: 30%, Testing: 30% Repeated for four random trails | 93.61 ± 0.58 | 5.49 ± 0.30 | 95.07 ± 0.45 | 737 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, V.; Bhavsar, A. Feature Importance for Human Epithelial (HEp-2) Cell Image Classification. J. Imaging 2018, 4, 46. https://doi.org/10.3390/jimaging4030046
Gupta V, Bhavsar A. Feature Importance for Human Epithelial (HEp-2) Cell Image Classification. Journal of Imaging. 2018; 4(3):46. https://doi.org/10.3390/jimaging4030046
Chicago/Turabian StyleGupta, Vibha, and Arnav Bhavsar. 2018. "Feature Importance for Human Epithelial (HEp-2) Cell Image Classification" Journal of Imaging 4, no. 3: 46. https://doi.org/10.3390/jimaging4030046




